nwalign
Globally align two sequences using Needleman-Wunsch algorithm
Syntax
Description
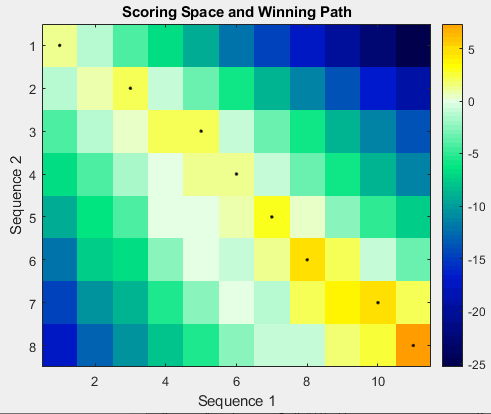
Score = nwalign(Seq1,Seq2)Seq1 and Seq2. The scale factor used to
calculate the score is provided by ScoringMatrix.
Score = nwalign(Seq1,Seq2,Name=Value)
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Durbin, Richard, Sean R. Eddy, Anders Krogh, and Graeme Mitchison. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. 1st ed. Cambridge University Press, 1998.
Version History
Introduced before R2006a
See Also
aa2int | aminolookup | baselookup | blosum | dayhoff | gonnet | int2aa | int2nt | localalign | multialign | nt2aa | nt2int | nuc44 | pam | profalign | seqdotplot | swalign