resubMargin
Resubstitution classification margins for multiclass error-correcting output codes (ECOC) model
Description
m = resubMargin(Mdl)m) for the multiclass error-correcting output codes (ECOC) model
Mdl using the training data stored in Mdl.X and
the corresponding class labels stored in Mdl.Y.
m is returned as a numeric column vector with the same length as
Mdl.Y. The software estimates each entry of m
using the trained ECOC model Mdl, the corresponding row of
Mdl.X, and the true class label Mdl.Y.
m = resubMargin(Mdl,Name,Value)
Examples
Calculate the resubstitution classification margins for an ECOC model with SVM binary learners.
Load Fisher's iris data set. Specify the predictor data X and the response data Y.
load fisheriris
X = meas;
Y = species;Train an ECOC model using SVM binary classifiers. Standardize the predictors using an SVM template, and specify the class order.
t = templateSVM('Standardize',true);
classOrder = unique(Y)classOrder = 3×1 cell
{'setosa' }
{'versicolor'}
{'virginica' }
Mdl = fitcecoc(X,Y,'Learners',t,'ClassNames',classOrder);
t is an SVM template object. During training, the software uses default values for empty properties in t. Mdl is a ClassificationECOC model.
Calculate the classification margins for the observations used to train Mdl. Display the distribution of the margins using a box plot.
m = resubMargin(Mdl);
boxplot(m)
title('In-Sample Margins')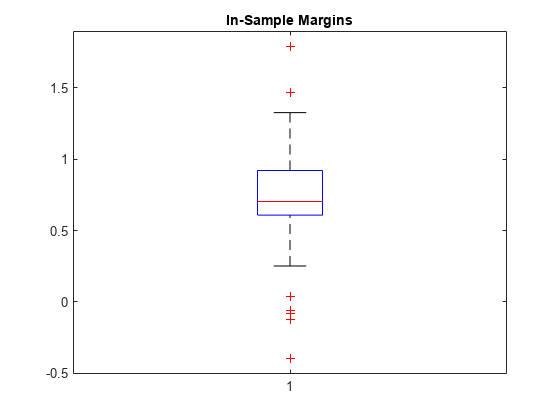
The classification margin of an observation is the positive-class negated loss minus the maximum negative-class negated loss. Choose classifiers that yield relatively large margins.
Perform feature selection by comparing training-sample margins from multiple models. Based solely on this comparison, the model with the greatest margins is the best model.
Load Fisher's iris data set. Define two data sets:
fullXcontains all four predictors.partXcontains the sepal measurements only.
load fisheriris
X = meas;
fullX = X;
partX = X(:,1:2);
Y = species;Train an ECOC model using SVM binary learners for each predictor set. Standardize the predictors using an SVM template, specify the class order, and compute posterior probabilities.
t = templateSVM('Standardize',true);
classOrder = unique(Y)classOrder = 3×1 cell
{'setosa' }
{'versicolor'}
{'virginica' }
FullMdl = fitcecoc(fullX,Y,'Learners',t,'ClassNames',classOrder,... 'FitPosterior',true); PartMdl = fitcecoc(partX,Y,'Learners',t,'ClassNames',classOrder,... 'FitPosterior',true);
Compute the resubstitution margins for each classifier. For each model, display the distribution of the margins using a boxplot.
fullMargins = resubMargin(FullMdl); partMargins = resubMargin(PartMdl); boxplot([fullMargins partMargins],'Labels',{'All Predictors','Two Predictors'}) title('Training-Sample Margins')
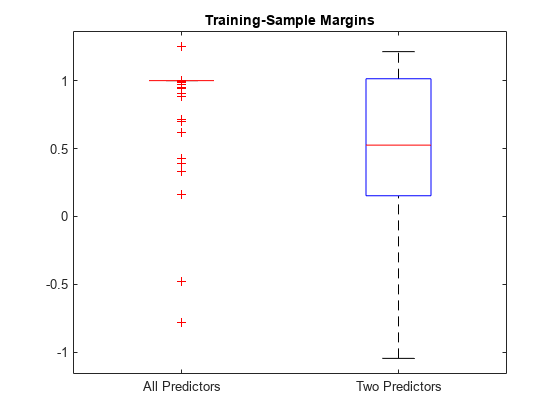
The margin distribution of FullMdl is situated higher and has less variability than the margin distribution of PartMdl. This result suggests that the model trained with all the predictors fits the training data better.
Input Arguments
Full, trained multiclass ECOC model, specified as a ClassificationECOC model trained with fitcecoc.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: resubMargin(Mdl,'Verbose',1) specifies to display diagnostic
messages in the Command Window.
Binary learner loss function, specified as a built-in loss function name or function handle.
This table describes the built-in functions, where yj is the class label for a particular binary learner (in the set {–1,1,0}), sj is the score for observation j, and g(yj,sj) is the binary loss formula.
Value Description Score Domain g(yj,sj) "binodeviance"Binomial deviance (–∞,∞) log[1 + exp(–2yjsj)]/[2log(2)] "exponential"Exponential (–∞,∞) exp(–yjsj)/2 "hamming"Hamming [0,1] or (–∞,∞) [1 – sign(yjsj)]/2 "hinge"Hinge (–∞,∞) max(0,1 – yjsj)/2 "linear"Linear (–∞,∞) (1 – yjsj)/2 "logit"Logistic (–∞,∞) log[1 + exp(–yjsj)]/[2log(2)] "quadratic"Quadratic [0,1] [1 – yj(2sj – 1)]2/2 The software normalizes binary losses so that the loss is 0.5 when yj = 0. Also, the software calculates the mean binary loss for each class [1].
For a custom binary loss function, for example
customFunction, specify its function handleBinaryLoss=@customFunction.customFunctionhas this form:bLoss = customFunction(M,s)
Mis the K-by-B coding matrix stored inMdl.CodingMatrix.sis the 1-by-B row vector of classification scores.bLossis the classification loss. This scalar aggregates the binary losses for every learner in a particular class. For example, you can use the mean binary loss to aggregate the loss over the learners for each class.K is the number of classes.
B is the number of binary learners.
For an example of passing a custom binary loss function, see Predict Test-Sample Labels of ECOC Model Using Custom Binary Loss Function.
This table identifies the default BinaryLoss value, which depends on the
score ranges returned by the binary learners.
| Assumption | Default Value |
|---|---|
All binary learners are any of the following:
| "quadratic" |
| All binary learners are SVMs or linear or kernel classification models of SVM learners. | "hinge" |
All binary learners are ensembles trained by
AdaboostM1 or
GentleBoost. | "exponential" |
All binary learners are ensembles trained by
LogitBoost. | "binodeviance" |
You specify to predict class posterior probabilities by setting
FitPosterior=true in fitcecoc. | "quadratic" |
| Binary learners are heterogeneous and use different loss functions. | "hamming" |
To check the default value, use dot notation to display the BinaryLoss property of the trained model at the command line.
Example: BinaryLoss="binodeviance"
Data Types: char | string | function_handle
Decoding scheme that aggregates the binary losses, specified as
"lossweighted" or "lossbased". For more
information, see Binary Loss.
Example: Decoding="lossbased"
Data Types: char | string
Estimation options, specified as a structure array as returned by statset.
To invoke parallel computing you need a Parallel Computing Toolbox™ license.
Example: Options=statset(UseParallel=true)
Data Types: struct
Verbosity level, specified as 0 or 1.
Verbose controls the number of diagnostic messages that the
software displays in the Command Window.
If Verbose is 0, then the software does not display
diagnostic messages. Otherwise, the software displays diagnostic messages.
Example: Verbose=1
Data Types: single | double
More About
The classification margin is, for each observation, the difference between the negative loss for the true class and the maximal negative loss among the false classes. If the margins are on the same scale, then they serve as a classification confidence measure. Among multiple classifiers, those that yield greater margins are better.
The binary loss is a function of the class and classification score that determines how well a binary learner classifies an observation into the class. The decoding scheme of an ECOC model specifies how the software aggregates the binary losses and determines the predicted class for each observation.
Assume the following:
mkj is element (k,j) of the coding design matrix M—that is, the code corresponding to class k of binary learner j. M is a K-by-B matrix, where K is the number of classes, and B is the number of binary learners.
sj is the score of binary learner j for an observation.
g is the binary loss function.
is the predicted class for the observation.
The software supports two decoding schemes:
Loss-based decoding [2] (
Decodingis"lossbased") — The predicted class of an observation corresponds to the class that produces the minimum average of the binary losses over all binary learners.Loss-weighted decoding [3] (
Decodingis"lossweighted") — The predicted class of an observation corresponds to the class that produces the minimum average of the binary losses over the binary learners for the corresponding class.The denominator corresponds to the number of binary learners for class k. [1] suggests that loss-weighted decoding improves classification accuracy by keeping loss values for all classes in the same dynamic range.
The predict, resubPredict, and
kfoldPredict functions return the negated value of the objective
function of argmin as the second output argument
(NegLoss) for each observation and class.
This table summarizes the supported binary loss functions, where yj is a class label for a particular binary learner (in the set {–1,1,0}), sj is the score for observation j, and g(yj,sj) is the binary loss function.
| Value | Description | Score Domain | g(yj,sj) |
|---|---|---|---|
"binodeviance" | Binomial deviance | (–∞,∞) | log[1 + exp(–2yjsj)]/[2log(2)] |
"exponential" | Exponential | (–∞,∞) | exp(–yjsj)/2 |
"hamming" | Hamming | [0,1] or (–∞,∞) | [1 – sign(yjsj)]/2 |
"hinge" | Hinge | (–∞,∞) | max(0,1 – yjsj)/2 |
"linear" | Linear | (–∞,∞) | (1 – yjsj)/2 |
"logit" | Logistic | (–∞,∞) | log[1 + exp(–yjsj)]/[2log(2)] |
"quadratic" | Quadratic | [0,1] | [1 – yj(2sj – 1)]2/2 |
The software normalizes binary losses so that the loss is 0.5 when yj = 0, and aggregates using the average of the binary learners [1].
Do not confuse the binary loss with the overall classification loss (specified by the
LossFun name-value argument of the resubLoss and
resubPredict object functions), which measures how well an ECOC
classifier performs as a whole.
Tips
To compare the margins or edges of several ECOC classifiers, use template objects to specify a common score transform function among the classifiers during training.
References
[1] Allwein, E., R. Schapire, and Y. Singer. “Reducing multiclass to binary: A unifying approach for margin classifiers.” Journal of Machine Learning Research. Vol. 1, 2000, pp. 113–141.
[2] Escalera, S., O. Pujol, and P. Radeva. “Separability of ternary codes for sparse designs of error-correcting output codes.” Pattern Recog. Lett. Vol. 30, Issue 3, 2009, pp. 285–297.
[3] Escalera, S., O. Pujol, and P. Radeva. “On the decoding process in ternary error-correcting output codes.” IEEE Transactions on Pattern Analysis and Machine Intelligence. Vol. 32, Issue 7, 2010, pp. 120–134.
Extended Capabilities
To run in parallel, specify the Options name-value argument in the call to
this function and set the UseParallel field of the
options structure to true using
statset:
Options=statset(UseParallel=true)
For more information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2014b
See Also
ClassificationECOC | resubEdge | margin | predict | resubPredict | fitcecoc | resubLoss
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)