2-D Discrete Wavelet Analysis
This section takes you through the features of 2-D discrete wavelet analysis using the Wavelet Toolbox™ software. The toolbox provides these functions for image analysis. For more information, see the function reference pages.
Note
In this section the presentation and examples use 2-D arrays corresponding to
indexed image representations. However, the functions described are also available
when using truecolor images, which are represented by
m-by-n-by-3 arrays of
uint8.
Analysis-Decomposition Functions
Synthesis-Reconstruction Functions
Decomposition Structure Utilities
Denoising and Compression
Function Name | Purpose |
|---|---|
Wavelet image denoising | |
Provide default values for denoising and compression | |
Penalized threshold for wavelet 1-D or 2-D denoising | |
Thresholds for wavelet 2-D using Birgé-Massart strategy | |
Wavelet denoising and compression | |
Threshold settings manager |
In this section, you'll learn
How to load an image
How to analyze an image
How to compress an image
Wavelet Image Analysis and Compression
This example shows how you can use 2-D wavelet analysis to compress an image efficiently without sacrificing its clarity.
Note: Instead of directly using image(I) to visualize the image I, we use image(wcodemat(I)), which displays a rescaled version of I leading to a clearer presentation of the details and approximations (see wcodemat).
Load an image.
load porche whos X map
Name Size Bytes Class Attributes X 512x512 2097152 double map 235x3 5640 double
Display the image.
image(X) colormap(map) colorbar
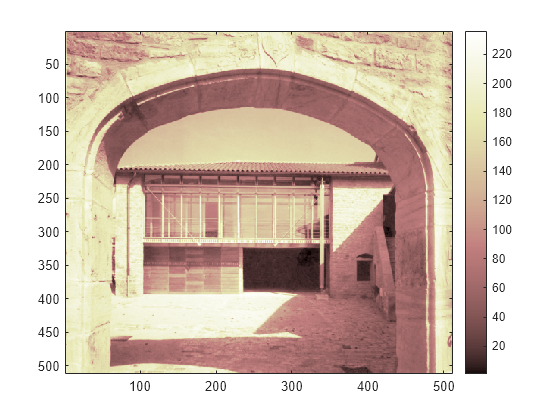
If the colormap is smooth, the wavelet transform can be directly applied to the indexed image; otherwise the indexed image should be converted to grayscale format. For more information, see Wavelets: Working with Images. Since the colormap is smooth in this image, you can now perform the decomposition.
Perform a single-level wavelet decomposition of the image using the bior3.7 wavelet. The coefficient matrix cA1 are the approximation coefficients. The horizontal, vertical, and diagonal details are in the matrices cH1, cV1, and cD1, respectively.
wv = 'bior3.7';
[cA1,cH1,cV1,cD1] = dwt2(X,wv);Use idwt1 to construct the approximations and details from the coefficients. (Note: You can also use upcoef2.)
sx = size(X); A1 = idwt2(cA1,[],[],[],wv,sx); H1 = idwt2([],cH1,[],[],wv,sx); V1 = idwt2([],[],cV1,[],wv,sx); D1 = idwt2([],[],[],cD1,wv,sx);
Display the approximations and details.
figure tiledlayout(2,2) nexttile image(wcodemat(A1,192)) title('Approximation A1') nexttile image(wcodemat(H1,192)) title('Horizontal Detail H1') nexttile image(wcodemat(V1,192)) title('Vertical Detail V1') nexttile image(wcodemat(D1,192)) title('Diagonal Detail D1') colormap(map)
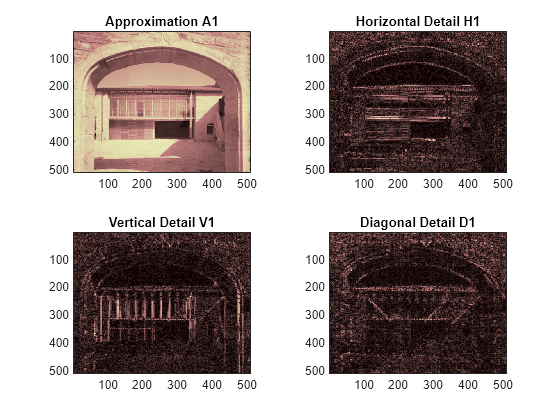
Regenerate the image by the single-level inverse discrete wavelet transform. Confirm the difference between the regenerated and original images are small.
Xrec = idwt2(cA1,cH1,cV1,cD1,wv); max(abs(X(:)-Xrec(:)))
ans = 1.9895e-13
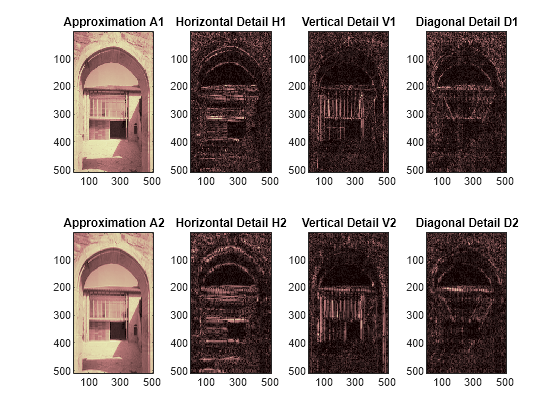
Perform a level-2 wavelet decomposition of the image using the same bior3.7 wavelet. The coefficients of all the components of a second-level decomposition (that is, the second-level approximation and the first two levels of detail) are returned concatenated into one vector, C. Argument S is a bookkeeping matrix that keeps track of the sizes of each component.
[c,s] = wavedec2(X,2,wv);
Extract the level 2 approximation coefficients. Extract the first- and second-level detail coefficients.
cA2 = appcoef2(c,s,wv,2); [cH2,cV2,cD2] = detcoef2('all',c,s,2); [cH1,cV1,cD1] = detcoef2('all',c,s,1);
Reconstruct the level 2 approximation and the level 1 and level 2 details.
A2 = wrcoef2('a',c,s,wv,2); H1 = wrcoef2('h',c,s,wv,1); V1 = wrcoef2('v',c,s,wv,1); D1 = wrcoef2('d',c,s,wv,1); H2 = wrcoef2('h',c,s,wv,2); V2 = wrcoef2('v',c,s,wv,2); D2 = wrcoef2('d',c,s,wv,2);
Display the approximation and details.
figure tiledlayout(2,4) nexttile image(wcodemat(A1,192)) title('Approximation A1') nexttile image(wcodemat(H1,192)) title('Horizontal Detail H1') nexttile image(wcodemat(V1,192)) title('Vertical Detail V1') nexttile image(wcodemat(D1,192)) title('Diagonal Detail D1') nexttile image(wcodemat(A2,192)) title('Approximation A2') nexttile image(wcodemat(H2,192)) title('Horizontal Detail H2') nexttile image(wcodemat(V2,192)) title('Vertical Detail V2') nexttile image(wcodemat(D2,192)) title('Diagonal Detail D2') colormap(map)
Compress the image. Use ddencmp to calculate the default parameters and wdencmp to perform the actual compression.
[thr,sorh,keepapp] = ddencmp('cmp','wv',X); [Xcomp,CXC,LXC,PERF0,PERFL2] = ... wdencmp('gbl',c,s,wv,2,thr,sorh,keepapp);
Compare the compressed image with the original image.
fprintf('Percentage of wavelet coefficients set to zero: %.4f\n',PERF0);Percentage of wavelet coefficients set to zero: 52.2313
fprintf('Percentage of energy preserved: %.4f\n',PERFL2);Percentage of energy preserved: 99.9982
figure tiledlayout(1,2) nexttile image(X) title('Original Image') axis square nexttile image(Xcomp) title('Compressed Image') axis square colormap(map)
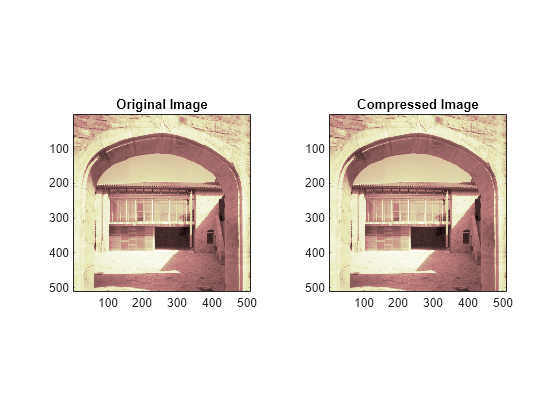
Note that, even though the compressed image is constructed from only about half as many nonzero wavelet coefficients as the original, there is almost no detectable deterioration in the image quality.