Results for
Cody has a wealth of problem groups that allow users of various skill levels to improve programming skills vis-à-vis MATLAB in an engaging way.
I would like to highlight the Draw Letters group, composed of problems created by Majid Farzaneh.

If you haven't yet visited Cody or solved that problem group, I would recommend that you head over there now. What are you waiting for?
Hi, all friends
I am dealing with a confusing NCA result. As this result shows, AUC_infinity=0.8537 (μg*h/mL), DM=1 (mg). CL should be equal to DM/AUC_infinity but why is the calculated result 2.4491? There is no dimension along with value so I do not know if I ignore anything.
Does any one can handle with this situation? Thanks very much. I also post my used data (i.v.).
Hi, all friends
If I define a parameter, like solubility with dimension of milligram/milliliter and I want to refer it in an reaction equation in dimension of millimole/liter, suppose I have define its molecularweight, could I input this parameter directly in equation with specified dimension?
Question: How to load a host of variants created through scripts to a sbproj file so that we can simulate and visualize them through the simbiology GUI.
Description : In a typical QSP workflow, we generate virtual patients by perturbing a set of parameters. These virtual patients are generated as variants by using "addvariant".
We end up with an array of variants saved as "variants.mat". This mat contains the list of ~3000 variants.
These variants are now to be simulated with different dose objects. This could be done with scripts but it would be ideal if we import these variants into the sbproj file and simulate via simbiology GUI for both purpose of troubleshooting and dosing.
Is there a way to do this?
We have created a new distance learning community for educators who are teaching remotely or online using MathWorks tools. It houses resources, such as articles, code examples, and videos, as well as an area where community members can ask questions or hold discussions around best practices in distance learning. Jiro Doke , who also writes for the File Exchange Pick of the Week blog, will be moderating this community.
We encourage you to visit the new community and share best practices, examples, and ask questions.
We have created a new page in MATLAB Answers that aggregates the most frequently asked MATLAB questions (FAQs). FAQs are common and a good starting point for anyone learning something new and having them all in one place can be a useful resource. Our new FAQ page lists the top 100 MATLAB questions asked and answered in MATLAB Central . They are grouped into the following 9 categories:
- Data Import and Analysis
- Graphics: Basics
- Graphics: Objects
- Installation and Licensing
- Language Fundamentals: Basics
- Language Fundamentals: Matrices and Arrays
- Language Fundamentals: Operators and Elementary Operations
- Mathematics
- Programming
I recommend that everyone scans the new FAQ page as you might find some useful tips.
We will be updating the FAQ page regularly as new questions are asked in the community. We welcome any feedback you have.
Hi, all friends
I am working on building a oral model with "Generic SimBiology PBPK model" and meet some problems about intestinal transit rate. Take duodenum as example, the transit rate is defined as " (kTransportSmallIntestine*organismLengthDuodenum/organismLengthSmallInstestine)*Duodenum.DrugDissolved". I think assuming duodenum transit time is equal to SmallIntestineTransittime*organismLengthDuodenum/organismLengthSmallInstestine, the transit rate constant is the inverse of that,which result in kTransportSmallIntestine*organismLengthSmallInstestine/organismLengthDuodenum, contradicting with the equation in model. Am I wrong?
Besides that, the kTransportSmallIntestine_is defined as _0.693/organismMeanResidenceSmallIntestine in model. Isn't the mean residence time determining the time at which about 63.2% of initial amount having passed through the compartment and inverse of mean residence time determining the transit rate? Why does organismMeanResidenceSmallIntestine correlate with 0.693 which is often seen in half-life associated expression?
Thanks for any comment.
Check out this pick of the week from Emma Gau. Emma's submission is featured on the Live Script gallery. Check out the blog post to see why Owen picked it.
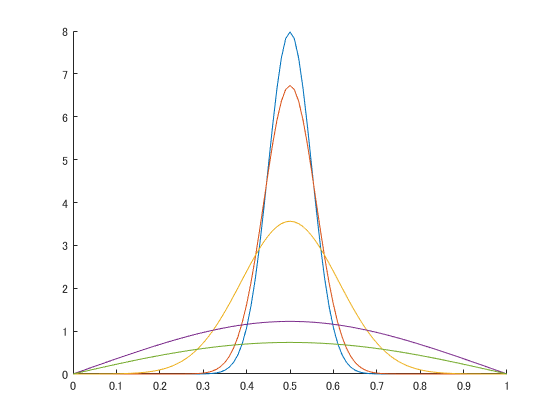
Hi, fellows,
I am a new user of MATLAB and SimBiology. When I open the "Generic SimBiology Physiologically-based Pharmacokinetic (PBPK) model" downloaded from "https://ww2.mathworks.cn/matlabcentral/fileexchange/37752-generic-simbiology-physiologically-based-pharmacokinetic-pbpk-model", I get a note of "Copyright 2012-2018 The MathWorks, Inc. ...". Does that mean I can not access to the model built-in or make any modification?
Thanks for any comment!
Check out this pick of the week from Will. Steve's progressbar submission has been around since 2005 and still runs perfectly. Check out the blog post to see why Will picked it.
We are excited to announce that Adam Danz has accepted our invitation and now is a member of the Community Advisory Board (CAB)!
Adam has been a rising star in Answers, obtaining 4500+ reputation points in the past year! Furthermore, he has contributed high-quality files to File Exchange, with an average rating of 4.8. Adam also demonstrates good communication skills and the ability to work with others. Those characteristics are what we expect to see from a CAB advisor. You can learn more about him and CAB on the CAB page .
On behalf of all the community team, we would like to extend our warmest welcome to Adam!

Check out Sean's blog posts ( Intro , Authoring ) on Spider Plots. He's using spider_plot by Moses to create beautiful plots like this.
New Features
R2019b introduces the new SimBiology Model Analyzer which replaces Task Editor enabling you to do a lot more within the app.
For a detailed look into enhancements with Model Analyzer, we created a few short videos. Below is a list of the new features together with videos describing each feature.
- Programs: Compose SimBiology analysis programs from built-in analysis steps. Video: Simulating a model using SimBiology Model Analyzer
- Scenarios: Generate Samples to perform Monte Carlo simulations using various modeling and dosing conditions. Video: Generating Simulation Scenarios by Sampling Model Quantities Video: Sampling Parameters from Covariate Models
- Plotting and Datasheets: Plot multi-dimensional data slicing by categories such as response or scenarios. Visualize and manipulate raw external and analysis data using datasheets. Video: Stratifying Data for Visualization in SimBiology
Compatibility considerations with earlier versions of SimBiology
There are a few things that one needs to be aware of if planning to use R2019b with a SimBiology project created in older versions. If you load a project created pre-R2019b, the existing tasks are converted to equivalent programs. However, keep in mind the following for the current version of SimBiology Model Analyzer:
- Live Plots functionality of the Task Editor is not supported. If Live Plots in the Task Editor is turned on and shows a time plot of simulation results, then an equivalent time plot is added on import to the SimBiology Model Analyzer app with the main difference being that the time plot is updated and displayed after the program finishes (not while it is running).
- Previously available Calculate Conserved Cycles, Search Model(s), and Generate Report tasks are not supported.
- Any Group Simulation task is converted to a Scan program.
- Fit Data program does not support nlinfit as the estimation method. However, the command-line function sbiofit still supports nlinfit.
- To help with the transition to R2019b, when you open an existing project created in R2019a or earlier, the app creates a backup of the original file. The backup file has the release information suffixed to its name, filename_release.sbproj. For example, if you open an R2019a project named foo.sbproj in the app, the app creates a backup file called foo_R2019a.sbproj.
- Also, when you open and save a project, the app creates a backup file of the version prior to the saved version of the project. The backup file has the .bak extension (for example, foo.sbproj.bak).
See the complete list of general behavior change, compatibility considerations, and new features in the release notes.
The following is a list of updates and new features for MATLAB Central, including MATLAB Answers, File Exchange, Blogs, and Cody.
New Features
Community highlights channel released - See this post for more details.
Answers comment UI change - A minor improvement to the user interface when adding comments to questions and answers in MATLAB Answers. This change should make it easier to discern between commenting and adding a new answers to a question.

Answers rich text editor update - The rich text editor has been updated to the 19b version of the MATLAB Live Editor. This update will primarily help with syntax highlighting along with resolving a few other issues.
Answer pages translation option - Answers translations options added. A translation option for questions and answers content has been added for French, German, Italian, Spanish, and Chinese languages.
Hey everyone! I'm spotlighting Nikolaos Nikolaou today because of the sheer quantity of his Cody solutions over such a relatively short time span. Nikolaos has submitted 1,161 Cody solutions over the last 6 months of 2019, averaging 6.4 solutions per day. Achieving a Cody rank of 19 with a score of 18,820 and earning him a plethora of badges (including the Speed Demon badge of course). He's completed 24 problem groups and is well into most of the remaining 35.
Way to go Nikolaos!
Check out nextname by Stephen Cobeldick, this weeks pick by Jiro. Read Jiro's blog post about why he picked Stephen's submission this week.

Please post any comments to Jiro's blog post here.
