isanomaly
Syntax
Description
tf = isanomaly(Mdl,Tbl)Tbl using the OneClassSVM object
Mdl and returns the logical array tf, whose
elements are true when an anomaly is detected in the corresponding row of
Tbl. You must use this syntax if you create Mdl
by passing a table to the ocsvm
function.
tf = isanomaly(___,ScoreThreshold=scoreThreshold)isanomaly identifies observations with scores
above scoreThreshold as anomalies.
Examples
Create a OneClassSVM object for uncontaminated training observations by using the ocsvm function. Then detect novelties (anomalies in new data) by passing the object and the new data to the object function isanomaly.
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau to predict whether an individual makes over $50,000 per year.
load census1994census1994 contains the training data set adultdata and the test data set adulttest.
ocsvm does not use observations with missing values. Remove missing values in the data sets to reduce memory consumption and speed up training.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
Train a one-class SVM for adultdata. Assume that adultdata does not contain outliers. Specify StandardizeData as true to standardize the input data, and set KernelScale to "auto" to let the function select an appropriate kernel scale parameter using a heuristic procedure.
rng("default") % For reproducibility [Mdl,~,s] = ocsvm(adultdata,StandardizeData=true,KernelScale="auto");
Mdl is a OneClassSVM object. If you do not specify the ContaminationFraction name-value argument as a value greater than 0, then ocsvm treats all training observations as normal observations. The function sets the score threshold to the maximum score value. Display the threshold value.
Mdl.ScoreThreshold
ans = 0.0322
Find anomalies in adulttest by using the trained one-class SVM model. Because you specified StandardizeData=true when you trained the model, the isanomaly function standardizes the input data by using the predictor means and standard deviations of the training data stored in the Mu and Sigma properties, respectively.
[tf_test,s_test] = isanomaly(Mdl,adulttest);
The isanomaly function returns the anomaly indicators tf_test and scores s_test for adulttest. By default, isanomaly identifies observations with scores above the threshold (Mdl.ScoreThreshold) as anomalies.
Create histograms for the anomaly scores s and s_test. Create a vertical line at the threshold of the anomaly scores.
h1 = histogram(s,NumBins=50,Normalization="probability"); hold on h2 = histogram(s_test,h1.BinEdges,Normalization="probability"); xline(Mdl.ScoreThreshold,"r-",join(["Threshold" Mdl.ScoreThreshold])) h1.Parent.YScale = 'log'; h2.Parent.YScale = 'log'; legend("Training Data","Test Data",Location="north") hold off
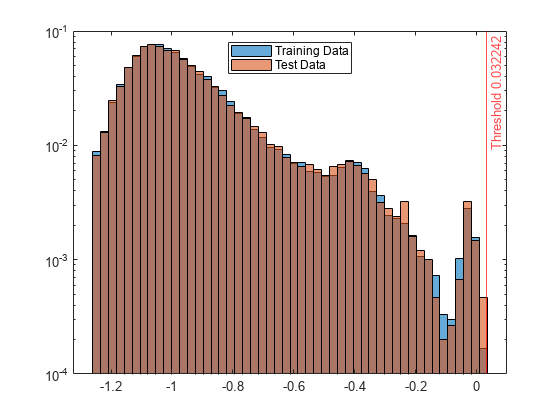
Display the observation index of the anomalies in the test data.
find(tf_test)
ans = 0×1 empty double column vector
The anomaly score distribution of the test data is similar to that of the training data, so isanomaly does not detect any anomalies in the test data with the default threshold value. You can specify a different threshold value by using the ScoreThreshold name-value argument. For an example, see Specify Anomaly Score Threshold.
Specify the threshold value for anomaly scores by using the ScoreThreshold name-value argument of isanomaly.
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau to predict whether an individual makes over $50,000 per year.
load census1994census1994 contains the training data set adultdata and the test data set adulttest.
ocsvm does not use observations with missing values. Remove missing values in the data sets to reduce memory consumption and speed up training.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
Train a one-class SVM model for adultdata. Specify StandardizeData as true to standardize the input data, and set KernelScale to "auto" to let the function select an appropriate kernel scale parameter using a heuristic procedure.
rng("default") % For reproducibility [Mdl,~,scores] = ocsvm(adultdata, ... StandardizeData=true,KernelScale="auto");
Plot a histogram of the score values. Create a vertical line at the default score threshold.
h = histogram(scores,NumBins=50,Normalization="probability"); h.Parent.YScale = 'log'; xline(Mdl.ScoreThreshold,"r-",join(["Threshold" Mdl.ScoreThreshold]))

Find the anomalies in the test data using the trained one-class SVM model. Use a different threshold from the default threshold value obtained when training the model.
First, determine the score threshold by using the isoutlier function.
[~,~,U] = isoutlier(scores)
U = -0.5342
Specify the value of the ScoreThreshold name-value argument as U. Because you specified StandardizeData as true when you trained Mdl, the isanomaly function standardizes new input data using the means and standard deviations stored in Mdl.Mu and Mdl.Sigma, respectively.
[tf_test,scores_test] = isanomaly(Mdl,adulttest,ScoreThreshold=U); h = histogram(scores_test,NumBins=50,Normalization="probability"); h.Parent.YScale = 'log'; xline(U,"r-",join(["Threshold" U]))

Input Arguments
Trained one-class SVM model, specified as a OneClassSVM
object.
Predictor data, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor variable.
Multicolumn variables and cell arrays other than cell arrays of character vectors are
not allowed.
If you train Mdl using a table, then you must provide predictor
data by using Tbl, not X. All predictor
variables in Tbl must have the same variable names and data types
as those in the training data. However, the column order in Tbl
does not need to correspond to the column order of the training data.
Data Types: table
Predictor data, specified as a numeric matrix. Each row of X
corresponds to one observation, and each column corresponds to one predictor
variable.
If you train Mdl using a matrix, then you must provide
predictor data by using X, not Tbl. The
variables that make up the columns of X must have the same order as
the training data.
Data Types: single | double
Threshold for the anomaly score, specified as a numeric scalar in the range
(–Inf,Inf). isanomaly identifies observations
with scores above the threshold as anomalies.
The default value is the ScoreThreshold property value of Mdl.
Example: ScoreThreshold=0.5
Data Types: single | double
Output Arguments
Anomaly indicators, returned as a logical column vector. An element of
tf is true when the observation in the
corresponding row of Tbl or X is an anomaly,
and false otherwise. tf has the same length as
Tbl or X.
isanomaly identifies observations with
scores above the threshold (the
scoreThreshold value) as anomalies.
Anomaly scores, returned as a numeric column vector whose values are in the range
(–Inf,Inf). scores has the same length as
Tbl or X, and each element of
scores contains an anomaly score for the observation in the
corresponding row of Tbl or X. A negative
score value with large magnitude indicates a normal observation, and a large positive
value indicates an anomaly.
Extended Capabilities
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for theisanomalyfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls theisanomalyfunction. Then usecodegento generate code for the entry-point function. For an example, see Code Generation for Anomaly Detection.To generate single-precision C/C++ code for
isanomaly, specifyDataType="single"when you call theloadLearnerForCoderfunction.Strict single-precision calculations are not supported. In the generated code, single-precision inputs produce single-precision outputs. However, variables inside the function might be double-precision.
This table contains notes about the arguments of
isanomaly. Arguments not included in this table are fully supported.Argument Notes and Limitations TblThe entry-point function must do the following:
Accept data as arrays.
Create a table from the data input arguments and specify the variable names in the table.
Pass the table to
isanomaly.
For an example of this table workflow, see Generate Code to Classify Data in Table. For more information on using tables in code generation, see Code Generation for Tables (MATLAB Coder) and Table Limitations for Code Generation (MATLAB Coder).
The number of rows, or observations, in
Tblcan be a variable size, but the number of columns inTblmust be fixed.
XThe number of rows, or observations, in
Xcan be a variable size, but the number of columns inXmust be fixed.ScoreThresholdNames in name-value arguments must be compile-time constants. UseParallelThis name-value argument is not supported, but the function supports parallel computation through OpenMP.
The generated code of
isanomalyusesparfor(MATLAB Coder) to create loops that run in parallel on supported shared-memory multicore platforms in the generated code. If your compiler does not support the Open Multiprocessing (OpenMP) application interface or you disable OpenMP library, MATLAB® Coder™ treats theparfor-loops asfor-loops. To find supported compilers, see Supported Compilers. To disable OpenMP library, set theEnableOpenMPproperty of the configuration object tofalse. For details, seecoder.CodeConfig(MATLAB Coder).
For more information, see Introduction to Code Generation.
isanomalyexecutes on a GPU if at least one of the following applies:The input argument
Xis agpuArrayobject.The input argument
TblcontainsgpuArrayvariables.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2022bisanomaly fully supports GPU arrays.
You can generate C/C++ code for the isanomaly function.
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)