OneClassSVM
Description
Use a one-class support vector machine model object OneClassSVM
for outlier detection and novelty detection.
Outlier detection (detecting anomalies in training data) — Detect anomalies in training data by using the
ocsvmfunction. Theocsvmfunction trains aOneClassSVMobject and returns anomaly indicators and scores for the training data.Novelty detection (detecting anomalies in new data with uncontaminated training data) — Create a
OneClassSVMobject by passing uncontaminated training data (data with no outliers) toocsvm, and detect anomalies in new data by passing the object and the new data to the object functionisanomaly. Theisanomalyfunction returns anomaly indicators and scores for the new data.
Creation
Create a OneClassSVM object by using the ocsvm
function.
Properties
This property is read-only.
Categorical predictor
indices, specified as a vector of positive integers. CategoricalPredictors
contains index values indicating that the corresponding predictors are categorical. The index
values are between 1 and p, where p is the number of
predictors used to train the model. If none of the predictors are categorical, then this
property is empty ([]).
This property is read-only.
Fraction of anomalies in the training data, specified as a numeric scalar in the
range [0,1].
If the
ContaminationFractionvalue is 0, thenocsvmtreats all training observations as normal observations, and sets the score threshold (ScoreThresholdproperty value) to the maximum anomaly score value of the training data.If the
ContaminationFractionvalue is in the range (0,1], thenocsvmdetermines the threshold value (ScoreThresholdproperty value) so that the function detects the specified fraction of training observations as anomalies.
This property is read-only.
Kernel scale parameter, specified as a positive scalar.
This property is read-only.
Regularization term strength, specified as a nonnegative scalar.
This property is read-only.
Predictor means of the training data, specified as a numeric vector.
If you specify
StandardizeData=truewhen you train a one-class SVM model usingocsvm:The length of
Muis equal to the number of predictors.If you set
StandardizeData=false, thenMuis an empty vector ([]).
This property is read-only.
Number of dimensions of the expanded space, specified as a positive integer.
This property is read-only.
Value of the objective function that the Limited-memory Broyden-Fletcher-Goldfarb-Shanno (LBFGS) solver minimizes to solve the one-class SVM problem, specified as a scalar.
This property is read-only.
Predictor variable names, specified as a cell array of character vectors. The order of the
elements in PredictorNames corresponds to the order in which the
predictor names appear in the training data.
This property is read-only.
Threshold for the anomaly score used to identify anomalies in the training data,
specified as a numeric scalar in the range (–Inf,Inf).
The software identifies observations with anomaly scores above the threshold as anomalies.
The
ocsvmfunction determines the threshold value to detect the specified fraction (ContaminationFractionproperty) of training observations as anomalies.
The
isanomalyobject function uses theScoreThresholdproperty value as the default value of theScoreThresholdname-value argument.
This property is read-only.
Predictor standard deviations of the training data, specified as a numeric vector.
If you specify
StandardizeData=truewhen you train a one-class SVM model usingocsvm:The length of
Sigmais equal to the number of predictors.If you set
StandardizeData=false, thenSigmais an empty vector ([]).
Object Functions
isanomaly | Find anomalies in data using one-class support vector machine (SVM) |
incrementalLearner | Convert one-class SVM model to incremental learner |
gather | Gather properties of Statistics and Machine Learning Toolbox object from GPU |
Examples
Detect outliers (anomalies in training data) by using the ocsvm function.
Load the sample data set NYCHousing2015.
load NYCHousing2015The data set includes 10 variables with information on the sales of properties in New York City in 2015. Display a summary of the data set.
summary(NYCHousing2015)
double
Values:
Min 1
Median 3
Max 5
NEIGHBORHOOD: 91446×1 cell array of character vectors
BUILDINGCLASSCATEGORY: 91446×1 cell array of character vectors
RESIDENTIALUNITS: 91446×1 double
Values:
Min 0
Median 1
Max 8759
COMMERCIALUNITS: 91446×1 double
Values:
Min 0
Median 0
Max 612
LANDSQUAREFEET: 91446×1 double
Values:
Min 0
Median 1700
Max 2.9306e+07
GROSSSQUAREFEET: 91446×1 double
Values:
Min 0
Median 1056
Max 8.9422e+06
YEARBUILT: 91446×1 double
Values:
Min 0
Median 1939
Max 2016
SALEPRICE: 91446×1 double
Values:
Min 0
Median 3.3333e+05
Max 4.1111e+09
SALEDATE: 91446×1 datetime
Values:
Min 01-Jan-2015
Median 09-Jul-2015
Max 31-Dec-2015
The SALEDATE column is a datetime array, which is not supported by ocsvm. Create columns for the month and day numbers of the datetime values, and delete the SALEDATE column.
[~,NYCHousing2015.MM,NYCHousing2015.DD] = ymd(NYCHousing2015.SALEDATE); NYCHousing2015.SALEDATE = [];
Train a one-class SVM model for NYCHousing2015. Specify the fraction of anomalies in the training observations as 0.1, and specify the first variable (BOROUGH) as a categorical predictor. The first variable is a numeric array, so ocsvm assumes it is a continuous variable unless you specify the variable as a categorical variable. In addition, specify StandardizeData as true to standardize the input data, because the predictors have largely different scales. Set KernelScale to "auto" so that the software selects an appropriate kernel scale parameter using a heuristic procedure.
rng("default") % For reproducibility [Mdl,tf,scores] = ocsvm(NYCHousing2015,ContaminationFraction=0.1, ... CategoricalPredictors=1,StandardizeData=true, ... KernelScale="auto");
Mdl is a OneClassSVM object. ocsvm also returns the anomaly indicators (tf) and anomaly scores (scores) for the training data NYCHousing2015.
Plot a histogram of the score values. Create a vertical line at the score threshold corresponding to the specified fraction.
histogram(scores) xline(Mdl.ScoreThreshold,"r-",["Threshold" Mdl.ScoreThreshold])
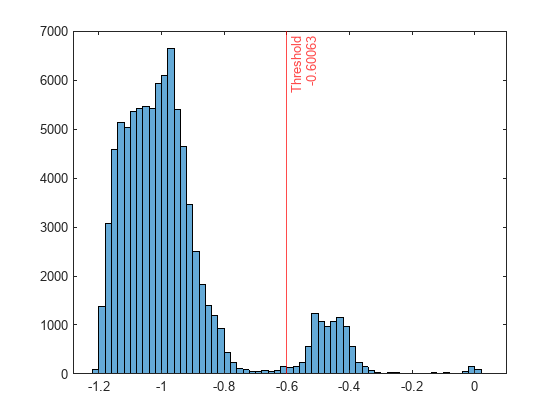
If you want to identify anomalies with a different contamination fraction (for example, 0.01), you can train a new one-class SVM model.
rng("default") % For reproducibility [newMdl,newtf,scores] = ocsvm(NYCHousing2015, ... ContaminationFraction=0.01,CategoricalPredictors=1, ... KernelScale="auto");
If you want to identify anomalies with a different score threshold value (for example, -0.7), you can pass the OneClassSVM object, the training data, and a new threshold value to the isanomaly function.
[newtf,scores] = isanomaly(Mdl,NYCHousing2015,ScoreThreshold=-0.7);
Note that changing the contamination fraction or score threshold changes the anomaly indicators only, and does not affect the anomaly scores. Therefore, if you do not want to compute the anomaly scores again by using ocsvm or isanomaly, you can obtain a new anomaly indicator with the existing score values.
Change the fraction of anomalies in the training data to 0.01.
newContaminationFraction = 0.01;
Find a new score threshold by using the quantile function.
newScoreThreshold = quantile(scores,1-newContaminationFraction)
newScoreThreshold = -0.3745
Obtain a new anomaly indicator.
newtf = scores > newScoreThreshold;
Create a OneClassSVM object for uncontaminated training observations by using the ocsvm function. Then detect novelties (anomalies in new data) by passing the object and the new data to the object function isanomaly.
Load the 1994 census data stored in census1994.mat. The data set consists of demographic data from the US Census Bureau to predict whether an individual makes over $50,000 per year.
load census1994census1994 contains the training data set adultdata and the test data set adulttest.
ocsvm does not use observations with missing values. Remove missing values in the data sets to reduce memory consumption and speed up training.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
Train a one-class SVM for adultdata. Assume that adultdata does not contain outliers. Specify StandardizeData as true to standardize the input data, and set KernelScale to "auto" to let the function select an appropriate kernel scale parameter using a heuristic procedure.
rng("default") % For reproducibility [Mdl,~,s] = ocsvm(adultdata,StandardizeData=true,KernelScale="auto");
Mdl is a OneClassSVM object. If you do not specify the ContaminationFraction name-value argument as a value greater than 0, then ocsvm treats all training observations as normal observations. The function sets the score threshold to the maximum score value. Display the threshold value.
Mdl.ScoreThreshold
ans = 0.0322
Find anomalies in adulttest by using the trained one-class SVM model. Because you specified StandardizeData=true when you trained the model, the isanomaly function standardizes the input data by using the predictor means and standard deviations of the training data stored in the Mu and Sigma properties, respectively.
[tf_test,s_test] = isanomaly(Mdl,adulttest);
The isanomaly function returns the anomaly indicators tf_test and scores s_test for adulttest. By default, isanomaly identifies observations with scores above the threshold (Mdl.ScoreThreshold) as anomalies.
Create histograms for the anomaly scores s and s_test. Create a vertical line at the threshold of the anomaly scores.
h1 = histogram(s,NumBins=50,Normalization="probability"); hold on h2 = histogram(s_test,h1.BinEdges,Normalization="probability"); xline(Mdl.ScoreThreshold,"r-",join(["Threshold" Mdl.ScoreThreshold])) h1.Parent.YScale = 'log'; h2.Parent.YScale = 'log'; legend("Training Data","Test Data",Location="north") hold off
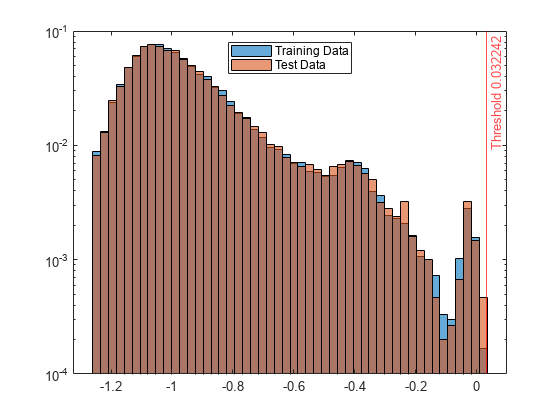
Display the observation index of the anomalies in the test data.
find(tf_test)
ans = 0×1 empty double column vector
The anomaly score distribution of the test data is similar to that of the training data, so isanomaly does not detect any anomalies in the test data with the default threshold value. You can specify a different threshold value by using the ScoreThreshold name-value argument. For an example, see Specify Anomaly Score Threshold.
More About
One-class SVM, or unsupervised SVM, is an
algorithm used for anomaly detection. The algorithm tries to separate data from the origin in
the transformed high-dimensional predictor space. ocsvm finds the decision
boundary based on the primal form of SVM with the Gaussian kernel approximation method.
Random feature expansion, such as Random Kitchen Sinks [1] or Fastfood [2], is a scheme to approximate Gaussian kernels of the kernel classification algorithm to use for big data in a computationally efficient way. Random feature expansion is more practical for big data applications that have large training sets, but can also be applied to smaller data sets that fit in memory.
The kernel classification algorithm searches for an optimal hyperplane that separates the data into two classes after mapping features into a high-dimensional space. Nonlinear features that are not linearly separable in a low-dimensional space can be separable in the expanded high-dimensional space. All the calculations for hyperplane classification use only dot products. You can obtain a nonlinear classification model by replacing the dot product x1x2' with the nonlinear kernel function , where xi is the ith observation (row vector) and φ(xi) is a transformation that maps xi to a high-dimensional space (called the “kernel trick”). However, evaluating G(x1,x2) (Gram matrix) for each pair of observations is computationally expensive for a large data set (large n).
The random feature expansion scheme finds a random transformation so that its dot product approximates the Gaussian kernel. That is,
where T(x) maps x in to a high-dimensional space (). The Random Kitchen Sinks scheme uses the random transformation
where is a sample drawn from and σ is a kernel scale. This scheme requires O(mp) computation and storage.
The Fastfood scheme introduces another random
basis V instead of Z using Hadamard matrices combined
with Gaussian scaling matrices. This random basis reduces the computation cost to O(mlogp) and reduces storage to O(m).
The ocsvm function uses the Fastfood
scheme for random feature expansion, and uses linear classification to train a one-class
Gaussian kernel classification model.
References
[1] Rahimi, A., and B. Recht. “Random Features for Large-Scale Kernel Machines.” Advances in Neural Information Processing Systems. Vol. 20, 2008, pp. 1177–1184.
[2] Le, Q., T. Sarlós, and A. Smola. “Fastfood — Approximating Kernel Expansions in Loglinear Time.” Proceedings of the 30th International Conference on Machine Learning. Vol. 28, No. 3, 2013, pp. 244–252.
Extended Capabilities
Usage notes and limitations:
The
isanomalyfunction supports code generation.
For more information, see Introduction to Code Generation.
Usage notes and limitations:
The following object functions fully support GPU arrays:
The object functions execute on a GPU if at least one of the following applies:
The model was fitted with GPU arrays.
The predictor data that you pass to the object function is a GPU array.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2022bYou can fit a OneClassSVM object with GPU arrays by using ocsvm. The
isanomaly object
function supports GPU array input arguments so that the function can execute on a GPU. The
incrementalLearner object function does not support GPU arrays.
You can also gather the properties of a OneClassSVM model object from
the GPU using the gather
function.
You can generate C/C++ code for the isanomaly function.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)