Classify Land Cover Using Sentinel-2 Multispectral Images
This example shows how to fine-tune a pretrained segmentation model backbone, using Sentinel-2 Level 2A multispectral images and CORINE Land Cover 2018 data, for pixel-level land-cover classification.
Sentinel-2 Level 2A data is multispectral data acquired in the Sentinel-2 mission. The Sentinel-2 mission under the Copernicus program [1] offers medium-resolution Earth imagery to track the variability in the conditions of the land surface of the Earth. The wide swath width and high revisit time of the mission facilitate the monitoring of changes on the surface of the Earth. Sentinel-2 Level 2A data contains 12 bands of image data having resolutions of 10 meters, 20 meters, and 60 meters. The size of a 10-meter resolution tile is 10,980-by-10,980 pixels. The CORINE Land Cover 2018 (vector), Europe, 6-yearly [2] data set consists of 44 thematic categories based on data from the year 2018.
This example first shows you how to perform semantic segmentation using a pretrained network to segment Sentinel-2 data based on land cover. Then, you can optionally fine-tune a pretrained network backbone on new Sentinel-2 data and CORINE Land Cover 2018 data using a patch-based training methodology. This example runs best on a CUDA® capable NVIDIA™ GPU. Use of a GPU requires Parallel Computing Toolbox™. For more information about the supported compute capabilities, see GPU Computing Requirements (Parallel Computing Toolbox).
Download Pretrained Network
Specify dataDir as the desired location of the pretrained network and data set.
dataDir = fullfile(tempdir,"sentinel2PretrainedModel");Download the pretrained network.
zipFile = matlab.internal.examples.downloadSupportFile("image","data/LandCoverClassificationModel.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
Load the pretrained network and class names into the workspace.
filename = fullfile(filepath,"LandCoverClassificationModel.mat"); load(filename,"net","classNames") numClasses = length(classNames);
Download Test Data
Create an account on the Copernicus Data Space website, and download a Sentinel-2 Level 2A tile as a zip file. The pretrained network has been trained on tiles representing regions in Europe, and likely works best on tiles in this region. However, you can use the pretrained network for any Sentinel-2 data. This example uses the tile S2B_MSIL2A_20181231T112459_N0211_R037_T29SND_20181231T122356.SAFE as test data for the pretrained network.
Extract a Sentinel-2 tile, and specify tileFolder as the full path and name of the extracted folder.
tileFolder =  "";
"";Load the tile size corresponding to a Sentinel-2 Level 2A resolution of 10 meters.
load(filename,"tileSize")Preprocess Data
Preprocess the Sentinel-2 data using the helperPreprocessSentinel2Cube helper function, attached to this example as a supporting file. The helper function performs these operations.
Resizes each band image to match the 10-meter resolution tile.
Concatenates all band images into a multispectral data cube of size 10,980-by-10,980-by-12.
Masks the pixel locations that have no data, corrupted data, cloud cover, or snow cover using masks from the scene classification layer (SCL) image that is part of the Sentinel-2 Level 2A data.
sentinel2Cube = helperPreprocessSentinel2Cube(tileFolder,tileSize);
Perform Semantic Segmentation
Use the pretrained network to perform semantic segmentation by using the predictSentinelTile helper function, attached to this example as a supporting file. The helper function performs semantic segmentation on the patches of the Sentinel-2 data cube using the pretrained network and generates a land cover mask. Save the results of the semantic segmentation.
pred = predictSentinelTile(sentinel2Cube,net); save("pred.mat","pred")
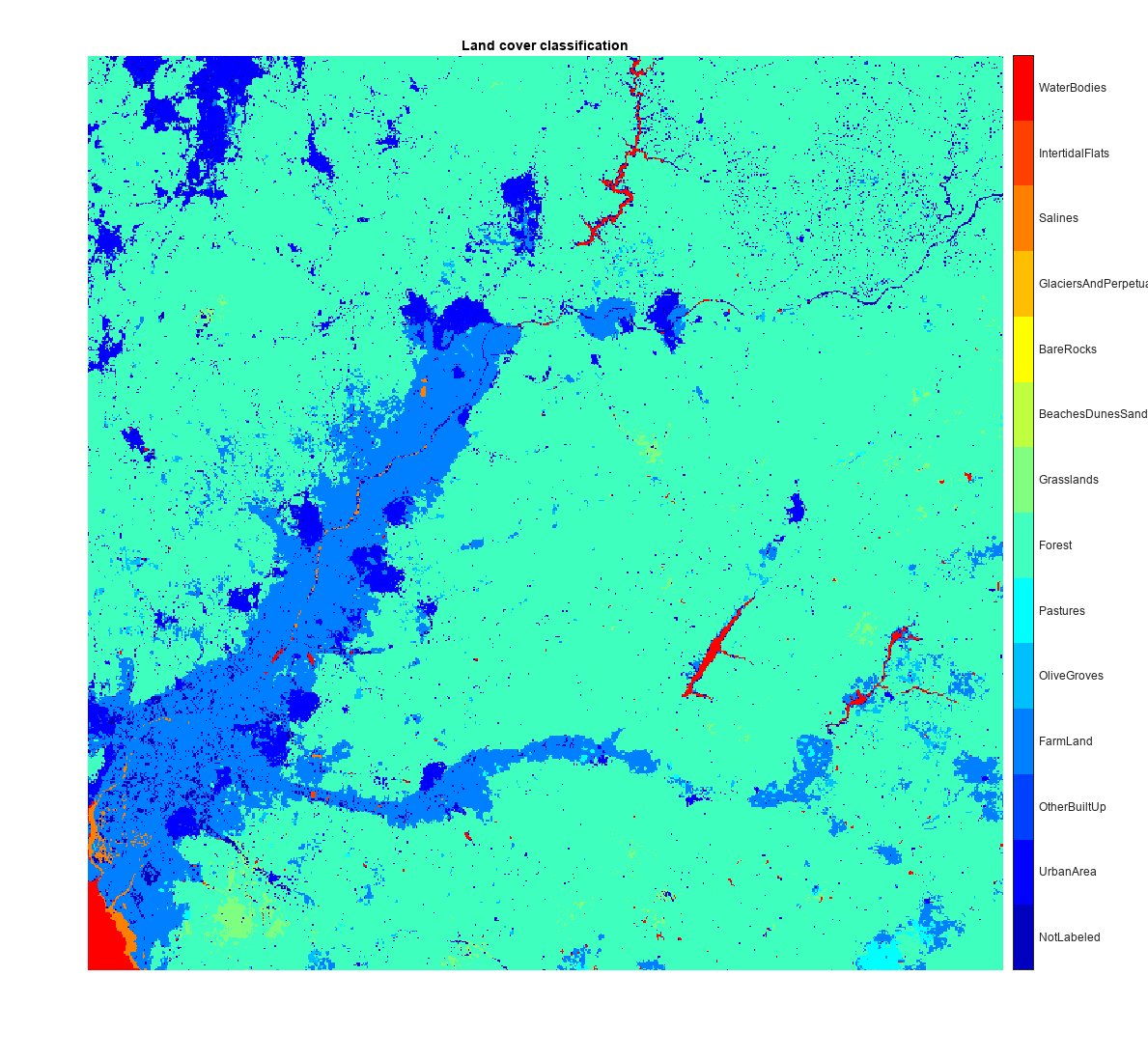
Visualize the truecolor image and the predicted land cover mask of the Sentinel-2 data.
TCI = dir(fullfile(tileFolder,"**"+filesep+"IMG_DATA"+filesep+"R10m"+filesep+"*_TCI*.jp2")); tciImage = imread(fullfile(TCI(1).folder,TCI(1).name)); figure imshow(tciImage) title("Truecolor Image")

ticks = 0.5:1:numClasses; cmap = jet(numClasses); figure imshow(pred,cmap) title("Land Cover Classification") colorbar(TickLabels=cellstr(classNames),Ticks=ticks,TickLength=0,TickLabelInterpreter="none")
The rest of this example shows how to fine-tune the pretrained network backbone using new Sentinel-2 Level 2A tiles and CORINE Land Cover 2018 data.
Download Sentinel-2 Data
If you do not already have an account on the Copernicus Data Space website, create one. Download some Sentinel-2 Level 2A tiles as zip files from the year 2018 from areas covered by CORINE Land Cover, which consists mostly of land regions within Europe. Ensure the data consists of various regions with different kinds of land cover, such as coastal regions, mountainous regions, urban and rural regions, and farm lands. This example uses these Sentinel-2 Level 2A tiles.
S2A_MSIL2A_20180812T124301_N0208_R095_T28WDT_20180812T154106.SAFES2A_MSIL2A_20181006T082811_N0208_R021_T36SWJ_20181006T104805.SAFES2A_MSIL2A_20181010T113321_N0209_R080_T30VVK_20181010T141708.SAFES2A_MSIL2A_20181024T111121_N0209_R137_T30STF_20181024T141739.SAFES2A_MSIL2A_20181117T105321_N0210_R051_T32ULE_20181117T121932.SAFES2A_MSIL2A_20181130T110411_N0211_R094_T30UYC_20181130T122430.SAFES2B_MSIL2A_20180303T125249_N9999_R138_T28VCR_20221029T232534S2B_MSIL2A_20181002T112109_N0208_R037_T29TNG_20181002T141523.SAFES2B_MSIL2A_20181006T110029_N0208_R094_T30SUG_20181006T154524.SAFES2B_MSIL2A_20181010T104019_N0206_R008_T32ULC_20181010T161145.SAFES2B_MSIL2A_20181029T111149_N0209_R137_T29SPB_20181029T140421.SAFES2B_MSIL2A_20181029T111149_N0209_R137_T29SQA_20181029T140421.SAFES2B_MSIL2A_20181102T123309_N0213_R052_T28WDS_20190903T160545.SAFES2B_MSIL2A_20181116T103309_N0210_R108_T32UME_20181116T152612.SAFES2B_MSIL2A_20181117T114349_N0210_R123_T30VUJ_20181117T143735.SAFES2B_MSIL2A_20181212T105439_N0211_R051_T31UES_20181218T134549.SAFES2B_MSIL2A_20181228T111449_N0211_R137_T30STG_20181228T134639.SAFES2B_MSIL2A_20181231T112459_N0211_R037_T29SND_20181231T122356.SAFES2B_MSIL2A_20181231T112459_N0211_R037_T29TNE_20181231T122356.SAFE
The latest version of the CORINE Land Cover is from the year 2018. Using Sentinel-2 data from that year is helpful for training, as the land cover labels match with the Sentinel-2 data. If the CORINE Land Cover releases new label data in the future, to use it for training, download Sentinel-2 tiles for that year.
Extract the Sentinel-2 tiles, and specify rootFolder as the full path and name of the extracted folder. If you are fine-tuning the network with the downloaded data for the first time, specify newData as true.
rootFolder =""; newData =
false;
Select a temporary directory to write the multispectral data cubes to. The specified directory must be on a drive with adequate disk space, approximately 1.34GB per downloaded tile. Preprocess and write each Sentinel-2 tile as a .tif file using the helperWriteCubes helper function, attached to this example as a supporting file. The helper function performs these operations.
Resizes each band image to match the 10-meter resolution tile.
Concatenates all band images into a multispectral data cube of size 10,980-by-10,980-by-12.
Masks the pixel locations that have no data, corrupted data, cloud cover, or snow cover using masks from the scene classification layer (SCL) image that is part of the Sentinel-2 Level 2A data.
Writes each Sentinel-2 tile as
.tiffiles.Returns the
RasterInfo(Mapping Toolbox) object of all tiles in a cell array.RasterInfoobjects contain information about geographic or projected raster data files, such as their file format, their native data type, and how they represent missing data.
tempDirForSentinel2CubeWrite =""; if newData Rinfo = helperWriteCubes(rootFolder,tempDirForSentinel2CubeWrite,tileSize); save("rinfo.mat","Rinfo") else load("rinfo.mat") end
Download the ground truth label data from the CORINE Land Cover website. Create an account, and download the "u2018_clc2018_v2020_20u1_fgdb" prepackaged data collection. Extract the downloaded labels, and specify europeCLClabelPath as the location of the geodatabase u2018_clc2018_v2020_20u1_fgdb\DATA\U2018_CLC2018_V2020_20u1.gdb.
europeCLClabelPath =  "";
"";Read the geodatabase containing the ground truth labels into a geospatial table using the readgeotable (Mapping Toolbox) function.
T = readgeotable(europeCLClabelPath);
Create Pixel Labels
Get the upper and lower bounds of each tile in world coordinates along the X and Y dimensions. This information is contained within the Rinfo variable returned by the helperWriteCubes helper function.
if newData lowerXWorldLimits = zeros(length(Rinfo),1); upperXWorldLimits = zeros(length(Rinfo),1); lowerYWorldLimits = zeros(length(Rinfo),1); upperYWorldLimits = zeros(length(Rinfo),1); for tileNumber = 1:length(Rinfo) lowerXWorldLimits(tileNumber) = Rinfo{tileNumber}.RasterReference.XWorldLimits(1); upperXWorldLimits(tileNumber) = Rinfo{tileNumber}.RasterReference.XWorldLimits(2); lowerYWorldLimits(tileNumber) = Rinfo{tileNumber}.RasterReference.YWorldLimits(1); upperYWorldLimits(tileNumber) = Rinfo{tileNumber}.RasterReference.YWorldLimits(2); end end
The Sentinel 2 tiles follow the WGS84 geographic coordinate reference system (CRS), while the CORINE Land Cover data follows the ETRS89-extended / LAEA Europe projected CRS. Hence, you must convert between these coordinate reference systems while generating labels. ETRS89-extended / LAEA Europe and WGS 84 are similar, but not identical. As a result, there can be slight discrepancies in coordinates between the two data.
The geodatabase table contains a row for each labeled region, each of which contains a mappolyshape (Mapping Toolbox) object and the label of the region. Search the regions in the geodatabase table that fall within the downloaded tiles and save them as a cell array of polyshape objects for each tile in the variable regionsWithinTile, along with their corresponding labels, in the variable numRegionsWithinTile. The geodatabase table contains more than 2.3 million entries, so this process might require significant processing time depending on your system configuration.
if newData regionsWithinTile = cell(length(Rinfo),1); regionLabel = cell(length(Rinfo),1); labels = unique(T.Code_18(1:end)); % Get unique labels T1 = geotable2table(T,["XWorld","YWorld"]); warning("off","MATLAB:polyshape:repairedBySimplify") % Turn off well-shaped polyshape warning for i = 1:height(T) % For each row in the geodatabase table, convert the mappolyshape to % XWorld and YWorld coordinates using geotable2table row = T1(i,:); XWorld = row.XWorld{1}; YWorld = row.YWorld{1}; if isempty(XWorld)||isempty(YWorld) continue end % Check if current labeled region appears within any of the selected tiles currentRegionLabel = find(row.Code_18 == labels); for tileNumber = 1:length(Rinfo) % Convert from ETRS89-extended / LAEA Europe to WGS84 [lat,lon] = projinv(T.Shape.ProjectedCRS,XWorld,YWorld); [x,y] = projfwd(Rinfo{tileNumber}.RasterReference.ProjectedCRS,lat,lon); roiPoly = [x(:) y(:)]; if any(roiPoly(:,1)>=lowerXWorldLimits(tileNumber) & roiPoly(:,1)<=upperXWorldLimits(tileNumber) & roiPoly(:,2)>=lowerYWorldLimits(tileNumber) & roiPoly(:,2)<=upperYWorldLimits(tileNumber)) % Add converted coordinates as a polyshape to the cell array of the tile in regionsWithinTile regionsWithinTile{tileNumber} = [regionsWithinTile{tileNumber} polyshape(roiPoly)]; regionLabel{tileNumber} = [regionLabel{tileNumber} currentRegionLabel]; end end end save("regionsWithinTile.mat","regionsWithinTile") save("regionLabel.mat","regionLabel") warning("on","MATLAB:polyshape:repairedBySimplify") % Turn on well-shaped polyshape warning else load("regionsWithinTile.mat") load("regionLabel.mat") end
Convert the generated polyshape lists to label masks for each tile and write them to the disk using the helperConvertPolyShapeToMask helper function. The helper function is attached to this example as a supporting file. Grouping labels of similar nature and spectral signature, such as different kinds of water bodies, into a single class improves model accuracy and reduces training time. Hence, group the 44 labels defined by the CORINE Land Cover data into 14 classes using the helperCombineLabels helper function. The helper function is attached to this example as a supporting file. To instead use all 44 original classes for training, skip this step.
To match the masking of cloud-covered and snow-covered regions and bad pixels performed on the Sentinel-2 data cubes during preprocessing, use the SCL image provided with the Sentinel-2 Level 2A data to mask the cloud-cover and snow-cover regions and bad pixels. For more information about the scene classification maps, see the Sentinel-2 L2A Scene Classification Map on the Sentinel Hub website.
tempDirForPixelLabelImageWrite =""; if newData for tileNumber = 1:length(Rinfo) mask = helperConvertPolyShapeToMask(regionsWithinTile{tileNumber},regionLabel1{tileNumber},tileSize,Rinfo{tileNumber}); mask = helperCombineLabels(mask); % Combine CORINE Land Cover labels SCL = dir(fullfile(rootFolder,"**"+filesep+"IMG_DATA"+filesep+"R20m"+filesep+"*_SCL*.jp2")); SCLimage = imresize(imread(fullfile(SCL(tileNumber).folder,SCL(tileNumber).name)),tileSize,"nearest"); SCLimage = SCLimage<1 | SCLimage>6; % Masking based on scene classification map legend mask(SCLimage) = 0; % Mask cloud-cover and snow-cover regions and pixels with data issues maskFilename = fullfile(tempDirForPixelLabelImageWrite,strcat("label_",erase(SCL(tileNumber).name,"_SCL_20m.jp2"))+".jpeg"); imwrite(mask,maskFilename,Mode="lossless",Quality=100) % Write image to disk end end
Prepare Data for Training
Prepare the data for training a DeepLab v3+ network based on ResNet-50. Different kinds of land cover occur in different proportions in different regions, creating an inevitable imbalance in the data set. Create matlab.io.datastore.FileSet objects of the data cube folder and the pixel label folder to help process a large collection of files.
fsImg = matlab.io.datastore.FileSet(tempDirForSentinel2CubeWrite); fsLab = matlab.io.datastore.FileSet(tempDirForPixelLabelImageWrite);
Specify two directories, one each in which to store the images and labels comprising the balanced data set.
imageDataset =""; labelDataset =
"";
Configure the pixel label balancing parameters. Specify 50,000 observations and a patch size corresponding to the size of the input of the network, which creates 50,000 images of patch size 224-by-224 and 12 bands and corresponding labels.
blockSize = [224 224];
numObservations =  50000;
50000;Create an array of blocked images from the written data.
blockedImageSentinel = blockedImage(fsImg); blockedImageLabels = blockedImage(fsLab);
Define class labels. These class names and labels are based on the grouping of classes created by the helperCombineLabels helper function.
classLabels = 0:length(classNames)-1;
Select block locations from the label images by using balancePixelLabels (Computer Vision Toolbox) to balance the classes.
locationSet = balancePixelLabels(blockedImageLabels,blockSize,numObservations,Classes=classNames,PixelLabelIDs=classLabels);
Write the patches of the data cube and labels in the balanced data set to disk. Sort locationSet by ImageNumber to write all patches from an image at one time, and then move to the next image. This helps reduce processing time by avoiding repeatedly reading from the same large data cubes. Although this induces a tile bias during training, you can counteract the bias by turning on shuffle in the model training options.
[s,idx] = sort(locationSet.ImageNumber); origins = locationSet.BlockOrigin(idx,:); imgNo = 0; for i = 1:length(s) if imgNo~=s(i) imgNo = imgNo + 1; img1 = imread(fsImg.FileInfo.Filename(imgNo)); img2 = imread(fsLab.FileInfo.Filename(imgNo)); end imgName = fullfile(imageDataset,join(string([origins(i,:),s(i)]),"_")+".tif"); block1img = img1(origins(i,1):origins(i,1)+blockSize(1)-1,origins(i,2):origins(i,2)+blockSize(2)-1,:); block2img = img2(origins(i,1):origins(i,1)+blockSize(1)-1,origins(i,2):origins(i,2)+blockSize(2)-1,:); noLabelRegionMask = (block2img == 0); noLabelRegionMask = repmat(noLabelRegionMask,1,1,size(block1img,3)); block1img(noLabelRegionMask) = 0; t = Tiff(imgName,"w"); tagstruct.ImageLength = blockSize(1); tagstruct.ImageWidth = blockSize(2); tagstruct.Photometric = Tiff.Photometric.MinIsBlack; tagstruct.BitsPerSample = 8; tagstruct.SamplesPerPixel = 12; tagstruct.RowsPerStrip = 16; tagstruct.PlanarConfiguration = Tiff.PlanarConfiguration.Chunky; tagstruct.Software = "MATLAB"; tagstruct.ExtraSamples = repmat(Tiff.ExtraSamples.Unspecified,[1,12-1]); setTag(t,tagstruct) write(t,block1img); close(t); labelName = fullfile(labelDataset,join(string([origins(i,:),s(i)]),"_")+".jpeg"); imwrite(block2img,labelName,Mode="lossless", Quality=100) end
Create an imageDatastore and a pixelLabelDatastore (Computer Vision Toolbox) for the patches of data cubes and labels, respectively.
imds = imageDatastore(imageDataset); pxds = pixelLabelDatastore(labelDataset,classNames,classLabels);
Randomly split the image and pixel label data into 60% training data, 20% validation data, and 20% test data using the partitionData helper function. The helper function is attached to this example as a supporting file.
[imdsTrain,imdsVal,imdsTest,pxdsTrain,pxdsVal,pxdsTest] = partitionData(imds,pxds);
Use data augmentation to improve network accuracy by randomly transforming the original data during training. Data augmentation adds more variety to the training data without increasing the number of labeled training samples. First, combine imdsTrain and pxdsTrain into a single CombinedDatastore.
dsTrain = combine(imdsTrain,pxdsTrain);
Then, apply the random transformation defined in the augmentImageAndPixelLabels helper function to both the image and pixel label training datastores. The helper function is attached to this example as a supporting file.
dsTrain = transform(dsTrain,@(dataIn)augmentImageAndPixelLabels(dataIn));
Combine the image and pixel label datastores of the validation data.
dsVal = combine(imdsVal,pxdsVal);
Define Network Architecture
Use the deeplabv3plus (Computer Vision Toolbox) function to create a DeepLab v3+ network based on ResNet-50. Modify the image input layer to support 12-channel data, instead of 3-channel data, and the first convolutional layer.
net = deeplabv3plus(blockSize,length(classNames),"resnet50"); layer = imageInputLayer([blockSize 12],Name="input_2",Normalization="none"); net = replaceLayer(net,"input_1",layer); layer = convolution2dLayer([7 7],64,Name="conv1",Padding=[3 3 3 3],Stride=[2 2]); net = replaceLayer(net,"conv1",layer);
Specify Training Options
Train the object detection network using the Adam optimization solver. Specify the hyperparameter settings using the trainingOptions (Deep Learning Toolbox) function. Specify the initial learning rate as 0.0001 and the mini-batch size as 16. You can experiment with tuning the hyperparameters based on your available system memory.
initialLearningRate = 0.0001; maxEpochs = 10; minibatchSize = 16; l2reg = 0.001; checkpointfolderloc =""; options = trainingOptions("adam", ... InitialLearnRate=initialLearningRate, ... L2Regularization=l2reg, ... MaxEpochs=maxEpochs, ... MiniBatchSize=minibatchSize, ... LearnRateSchedule="piecewise", ... LearnRateDropPeriod=1, ... LearnRateDropFactor=0.1, ... Shuffle="every-epoch", ... Plots="training-progress", ... VerboseFrequency=20, ... CheckpointPath=checkpointfolderloc, ... CheckpointFrequency=5, ... ExecutionEnvironment="gpu", ... ValidationData=dsVal, ... ValidationFrequency=100, ... OutputNetwork="best-validation-loss", ... ValidationPatience=100);
Train Network
To train the network, set the doTraining variable to true. Train the network using focal crossentropy loss on the available data. The loss function is defined in the modelLoss helper function. The helper function is attached to this example as a supporting file. Training this network for 50,000 observations takes about 6 hours on an NVIDIA™ GeForce RTX 4080 with 16 GB of GPU memory. Training may take longer depending on your GPU hardware and specified number of observations. If you have a GPU with less memory, lower the mini-batch size using the trainingOptions function, or reduce the network input size and resize the training data to avoid running out of memory.
doTraining = false; if doTraining net = trainnet(dsTrain,net,@modelLoss,options); save("LandCoverClassificationModel","net") else model = load("LandCoverClassificationModel"); net = model.net; end
Evaluate Network on Test Data
Select any of the Sentinel-2 whole tile data cubes written to path in the folder tempDirForSentinel2CubeWrite, and specify its index to the imgIndx variable. Predict the land cover for the entire data cube using the predictSentinelTile helper function. The helper function is attached to this example as a supporting file.
sentinel2Cubes = matlab.io.datastore.FileSet(tempDirForSentinel2CubeWrite);
gTruth = matlab.io.datastore.FileSet(tempDirForPixelLabelImageWrite);
imgIndx =  1;
pred = predictSentinelTile(imread(sentinel2Cubes.FileInfo.Filename(imgIndx)),net);
1;
pred = predictSentinelTile(imread(sentinel2Cubes.FileInfo.Filename(imgIndx)),net);Visualize the predicted and ground truth land cover classification labels.
montage({label2rgb(pred) label2rgb(imread(gTruth.FileInfo.Filename(imgIndx)))})
title("Model Prediction vs Ground Truth")
Predict the land cover classification for the test data set using the semanticseg (Computer Vision Toolbox) function. The semanticseg function returns the results for the test data set as a PixelLabelDatastore object and writes the actual pixel label data for each test image in imdsTest to the location specified by the WriteLocation name-value argument.
pxdsResults = semanticseg(imdsTest,net, ... MiniBatchSize=4, ... WriteLocation=tempdir, ... Verbose=false, ... Classes=classNames);
Measure semantic segmentation metrics on the test data set using the evaluateSemanticSegmentation (Computer Vision Toolbox) function.
metrics = evaluateSemanticSegmentation(pxdsResults,pxdsTest,Metrics=["global-accuracy","accuracy","weighted-iou"]);
Evaluating semantic segmentation results
----------------------------------------
* Selected metrics: global accuracy, class accuracy, weighted IoU.
* Processed 15983 images.
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy WeightedIoU
______________ ____________ ___________
0.84318 0.7143 0.743
Observe the high-level overview in the data set metrics by inspecting the DataSetMetrics property of the semanticSegmentationMetrics (Computer Vision Toolbox) object metrics. Observe the impact of each class on the overall performance and the per-class metrics by inspecting ClassMetrics property of metrics.
metrics.DataSetMetrics
ans=1×3 table
GlobalAccuracy MeanAccuracy WeightedIoU
______________ ____________ ___________
0.84318 0.7143 0.743
metrics.ClassMetrics
ans=14×1 table
Accuracy
________
NotLabeled 0.97319
UrbanArea 0.70758
OtherBuiltUp 0.2477
FarmLand 0.80356
OliveGroves 0.82728
Pastures 0.58053
Forest 0.77292
Grasslands 0.80955
BeachesDunesSands 0.42371
BareRocks 0.72372
GlaciersAndPerpetualSnow 0.67028
Salines 0.64909
IntertidalFlats 0.86539
WaterBodies 0.94571
The thematic accuracy of the CORINE land cover data is greater than or equal to 85% [2]. Considering the CORINE land cover data as a benchmark, the overall accuracy on the test data set is quite high. However, the class metrics show that underrepresented classes or classes with a lot of variety, such as OtherBuiltUp and BeachesDunesSands, are not segmented as well as classes such as Farmlands and Waterbodies. Additional data that includes more samples of the underrepresented classes and a use-case-specific grouping of data might improve the results.
References
[1] Ecosystem, Copernicus Data Space. “Copernicus Data Space Ecosystem | Europe’s Eyes on Earth,” June 21, 2024. https://dataspace.copernicus.eu/.
[2] “CORINE Land Cover 2018 (Vector/Raster 100 m), Europe, 6-Yearly.” https://land.copernicus.eu/en/products/corine-land-cover/clc2018.
[3] European Environment Agency and European Environment Agency. “CORINE Land Cover 2018 (Vector), Europe, 6-Yearly - Version 2020_20u1, May 2020.” FGeo,Spatialite. June 14, 2019. https://doi.org/10.2909/71C95A07-E296-44FC-B22B-415F42ACFDF0.
See Also
imageDatastore | pixelLabelDatastore (Computer Vision Toolbox) | deeplabv3plus (Computer Vision Toolbox) | trainingOptions (Deep Learning Toolbox) | semanticseg (Computer Vision Toolbox) | evaluateSemanticSegmentation (Computer Vision Toolbox)