lasso
Lasso or elastic net regularization for linear models
Description
B = lasso(X,y)X and the response y. Each
column of B corresponds to a particular regularization
coefficient in Lambda. By default, lasso
performs lasso regularization using a geometric sequence of
Lambda values.
B = lasso(X,y,Name=Value)Alpha=0.5 sets elastic
net as the regularization method, with the parameter Alpha equal
to 0.5.
Examples
Construct a data set with redundant predictors and identify those predictors by using lasso.
Create a matrix X of 100 five-dimensional normal variables. Create a response vector y from just two components of X, and add a small amount of noise.
rng default % For reproducibility X = randn(100,5); weights = [0;2;0;-3;0]; % Only two nonzero coefficients y = X*weights + randn(100,1)*0.1; % Small added noise
Construct the default lasso fit.
B = lasso(X,y);
Find the coefficient vector for the 25th Lambda value in B.
B(:,25)
ans = 5×1
0
1.6093
0
-2.5865
0
lasso identifies and removes the redundant predictors.
Create sample data with predictor variable X and response variable .
rng default % For reproducibility X = rand(100,1); y = 2*X + randn(100,1)/10;
Specify a regularization value, and find the coefficient of the regression model without an intercept term.
lambda = 1e-03; B = lasso(X,y,Lambda=lambda,Intercept=false)
Warning: When the 'Intercept' value is false, the 'Standardize' value is set to false.
B = 1.9825
Plot the real values (points) against the predicted values (line).
scatter(X,y) hold on x = 0:0.1:1; plot(x,x*B) hold off

Construct a data set with redundant predictors and identify those predictors by using cross-validated lasso.
Create a matrix X of 100 five-dimensional normal variables. Create a response vector y from two components of X, and add a small amount of noise.
rng default % For reproducibility X = randn(100,5); weights = [0;2;0;-3;0]; % Only two nonzero coefficients y = X*weights + randn(100,1)*0.1; % Small added noise
Construct the lasso fit by using 10-fold cross-validation with labeled predictor variables.
[B,FitInfo] = lasso(X,y,CV=10,PredictorNames=["x1","x2","x3","x4","x5"]);
Display the variables in the model that corresponds to the minimum cross-validated mean squared error (MSE).
idxLambdaMinMSE = FitInfo.IndexMinMSE; minMSEModelPredictors = FitInfo.PredictorNames(B(:,idxLambdaMinMSE)~=0)
minMSEModelPredictors = 1×2 cell
{'x2'} {'x4'}
Display the variables in the sparsest model within one standard error of the minimum MSE.
idxLambda1SE = FitInfo.Index1SE; sparseModelPredictors = FitInfo.PredictorNames(B(:,idxLambda1SE)~=0)
sparseModelPredictors = 1×2 cell
{'x2'} {'x4'}
In this example, lasso identifies the same predictors for the two models and removes the redundant predictors.
Visually examine the cross-validated error of various levels of regularization.
Load the sample data.
load acetyleneCreate a design matrix with interactions and no constant term.
X = [x1 x2 x3]; D = x2fx(X,"interaction"); D(:,1) = []; % No constant term
Construct the lasso fit using 10-fold cross-validation. Include the FitInfo output so you can plot the result.
rng default % For reproducibility [B,FitInfo] = lasso(D,y,CV=10);
Plot the cross-validated fits. The green circle and dotted line locate the Lambda with minimum cross-validation error. The blue circle and dotted line locate the point with minimum cross-validation error plus one standard error.
lassoPlot(B,FitInfo,PlotType="CV"); legend("show")

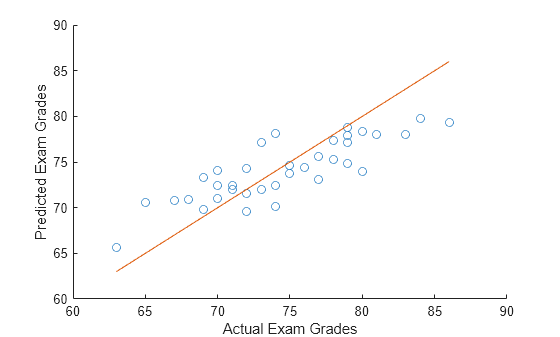
Predict students' exam scores using lasso and the elastic net method.
Load the examgrades data set.
load examgrades
X = grades(:,1:4);
y = grades(:,5);Split the data into training and test sets.
n = length(y); c = cvpartition(n,HoldOut=0.3); idxTrain = training(c,1); idxTest = ~idxTrain; XTrain = X(idxTrain,:); yTrain = y(idxTrain); XTest = X(idxTest,:); yTest = y(idxTest);
Find the coefficients of a regularized linear regression model using 10-fold cross-validation and the elastic net method with Alpha = 0.75. Use the largest Lambda value such that the mean squared error (MSE) is within one standard error of the minimum MSE.
[B,FitInfo] = lasso(XTrain,yTrain,Alpha=0.75,CV=10); idxLambda1SE = FitInfo.Index1SE; coef = B(:,idxLambda1SE); coef0 = FitInfo.Intercept(idxLambda1SE);
Predict exam scores for the test data. Compare the predicted values to the actual exam grades using a reference line.
yhat = XTest*coef + coef0; hold on scatter(yTest,yhat) plot(yTest,yTest) xlabel("Actual Exam Grades") ylabel("Predicted Exam Grades") hold off
Create a matrix X of N p-dimensional normal variables, where N is large and p = 1000. Create a response vector y from the model y = beta0 + X*p, where beta0 is a constant, along with additive noise.
rng default % For reproducibility N = 1e4; % Number of samples p = 1e3; % Number of features X = randn(N,p); beta = randn(p,1); % Multiplicative coefficients beta0 = randn; % Additive term y = beta0 + X*beta + randn(N,1); % Last term is noise
Construct the default lasso fit. Time the creation.
B = lasso(X,y,UseCovariance=false); % Warm up lasso for reliable timing data
tic
B = lasso(X,y,UseCovariance=false);
timefalse = toctimefalse = 8.9485
Construct the lasso fit using the covariance matrix. Time the creation.
B2 = lasso(X,y,UseCovariance=true); % Warm up lasso for reliable timing data
tic
B2 = lasso(X,y,UseCovariance=true);
timetrue = toctimetrue = 0.5156
The fitting time with the covariance matrix is much less than the time without it. View the speedup factor that results from using the covariance matrix.
speedup = timefalse/timetrue
speedup = 17.3553
Check that the returned coefficients B and B2 are similar.
norm(B-B2)/norm(B)
ans = 5.2508e-15
The results are virtually identical.
Input Arguments
Predictor data, specified as a numeric matrix. Each row represents one observation, and each column represents one predictor variable.
Data Types: single | double
Response data, specified as a numeric vector. y has
length n, where n is the number of
rows of X. The response y(i)
corresponds to the ith row of
X.
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: lasso(X,y,Alpha=0.75,CV=10) performs elastic net
regularization with 10-fold cross-validation. The Alpha=0.75
name-value pair argument sets the parameter used in the elastic net
optimization.
Absolute error tolerance used to determine the convergence of the
ADMM Algorithm, specified as a
positive scalar. The algorithm converges when successive estimates of
the coefficient vector differ by an amount less than
AbsTol.
Note
This option applies only when you use lasso
on tall arrays. See Extended Capabilities for more
information.
Example: AbsTol=1e–3
Data Types: single | double
Weight of lasso (L1)
versus ridge (L2)
optimization, specified as a positive scalar value in the interval
(0,1]. The value
Alpha = 1 represents lasso regression,
Alpha close to 0 approaches
ridge regression, and other
values represent elastic net optimization. See Elastic Net.
Example: Alpha=0.5
Data Types: single | double
Initial values for x-coefficients in ADMM Algorithm, specified as a numeric vector.
Note
This option applies only when you use lasso
on tall arrays. See Extended Capabilities for more
information.
Data Types: single | double
Size of the covariance matrix in megabytes, specified as a positive scalar or
"maximal". The lasso function can use
a covariance matrix for fitting when the UseCovariance argument is
true or "auto".
If UseCovariance is true or "auto"
and CacheSize is "maximal",
lasso can attempt to allocate a covariance matrix that
exceeds the available memory. In this case, MATLAB® issues an error.
Example: CacheSize="maximal"
Data Types: double | char | string
Cross-validation specification for estimating the mean squared error (MSE), specified as one of the following:
"resubstitution"—lassousesXandyto fit the model and to estimate the MSE without cross-validation.Positive scalar integer
K—lassousesK-fold cross-validation.cvpartitionobjectcvp—lassouses the cross-validation method expressed incvp. You cannot use a'leaveout'or custom'holdout'partition withlasso.
Example: CV=3
Maximum number of nonzero coefficients in the model, specified as a
positive integer scalar. lasso returns results only
for Lambda values that satisfy this
criterion.
Example: DFmax=5
Data Types: single | double
Flag for fitting the model with the intercept term, specified as
either true or false. The default
value is true, which indicates to include the
intercept term in the model. If Intercept is
false, then the returned intercept value is
0.
Example: Intercept=false
Data Types: logical
Regularization coefficients, specified as a vector of nonnegative values. See Lasso.
If you do not supply
Lambda, thenlassocalculates the largest value ofLambdathat gives a nonnull model. In this case,LambdaRatiogives the ratio of the smallest to the largest value of the sequence, andNumLambdagives the length of the vector.If you supply
Lambda, thenlassoignoresLambdaRatioandNumLambda.If
Standardizeistrue, thenLambdais the set of values used to fit the models with theXdata standardized to have zero mean and a variance of one.
The default is a geometric sequence of NumLambda
values, with only the largest value able to produce
B = 0.
Example: Lambda=linspace(0,1)
Data Types: single | double
Ratio of the smallest to the largest Lambda
values when you do not supply Lambda, specified as
a positive scalar.
If you set LambdaRatio = 0, then
lasso generates a default sequence of
Lambda values and replaces the smallest one
with 0.
Example: LambdaRatio=1e–2
Data Types: single | double
Maximum number of iterations allowed, specified as a positive integer scalar.
If the algorithm executes MaxIter iterations
before reaching the convergence tolerance RelTol,
then the function stops iterating and returns a warning message.
The function can return more than one warning when
NumLambda is greater than
1.
Default values are 1e5 for standard data and
1e4 for tall arrays.
Example: MaxIter=1e3
Data Types: single | double
Number of Monte Carlo repetitions for cross-validation, specified as a positive integer scalar.
If
CVis"resubstitution"or acvpartitionof type"resubstitution", thenMCRepsmust be1.If
CVis acvpartitionof type"holdout", thenMCRepsmust be greater than1.If
CVis a customcvpartitionof type"kfold", thenMCRepsmust be1.
Example: MCReps=5
Data Types: single | double
Options for computing in parallel and setting random streams, specified as a structure.
Parallel computation is not used when CV is
"resubstitution". Create the Options
structure using statset. This table lists the option
fields and their values.
| Field Name | Value | Default |
|---|---|---|
UseParallel | Set this value to true to run computations in
parallel. | false |
UseSubstreams | Set this value to To compute
reproducibly, set | false |
Streams | Specify this value as a RandStream object or
cell array of such objects. Use a single object except when the
UseParallel value is true
and the UseSubstreams value is
false. In that case, use a cell array that
has the same size as the parallel pool. | If you do not specify Streams, then
lasso uses the default stream or
streams. |
Note
You need Parallel Computing Toolbox™ to run computations in parallel.
Example: Options=statset(UseParallel=true,UseSubstreams=true,Streams=RandStream("mlfg6331_64"))
Data Types: struct
Names of the predictor variables, in the order in which they appear in
X, specified as a string array or cell array of
character vectors.
Example: PredictorNames=["x1","x2","x3","x4"]
Data Types: string | cell
Convergence threshold for the coordinate descent algorithm [3], specified as a positive scalar. The
algorithm terminates when successive estimates of the coefficient vector
differ in the L2 norm by a
relative amount less than RelTol.
Example: RelTol=5e–3
Data Types: single | double
Augmented Lagrangian parameter ρ for the ADMM Algorithm, specified as a positive scalar. The default is automatic selection.
Note
This option applies only when you use lasso
on tall arrays. See Extended Capabilities for more
information.
Example: Rho=2
Data Types: single | double
Flag for standardizing the predictor data X
before fitting the models, either true or
false. If Standardize is
true, then the X data is
scaled to have zero mean and a variance of one.
Standardize affects whether the regularization
is applied to the coefficients on the standardized scale or the original
scale. The results are always presented on the original data
scale.
If Intercept is false, then
the software sets Standardize to
false, regardless of the
Standardize value you specify.
X and y are always centered
when Intercept is true.
Example: Standardize=false
Data Types: logical
Indication to use a covariance matrix for fitting, specified as
"auto" or a logical scalar.
"auto"causeslassoto attempt to use a covariance matrix for fitting when the number of observations is greater than the number of problem variables. This attempt can fail when memory is insufficient. To find out whetherlassoused a covariance matrix for fitting, examine theUseCovariancefield of theFitInfooutput.truecauseslassoto use a covariance matrix for fitting as long as the required size does not exceedCacheSize. If the required covariance matrix size exceedsCacheSize,lassoissues a warning and does not use a covariance matrix for fitting.falsecauseslassonot to use a covariance matrix for fitting.
Using a covariance matrix for fitting can be faster than not using one, but can require more memory. See Use Correlation Matrix for Fitting Lasso. The speed increase can negatively affect numerical stability. For details, see Coordinate Descent Algorithm.
Example: UseCovariance=true
Data Types: logical | char | string
Initial value of the scaled dual variable u in the ADMM Algorithm, specified as a numeric vector.
Note
This option applies only when you use lasso
on tall arrays. See Extended Capabilities for more
information.
Data Types: single | double
Observation weights, specified as a nonnegative vector.
Weights has length n, where
n is the number of rows of
X. The lasso function scales
Weights to sum to 1.
Data Types: single | double
Output Arguments
Fitted coefficients, returned as a numeric matrix. B
is a p-by-L matrix, where
p is the number of predictors (columns) in
X, and L is the number of
Lambda values. You can specify the number of
Lambda values using the
NumLambda name-value pair argument.
The coefficient corresponding to the intercept term is a field in
FitInfo.
Data Types: single | double
Fit information of the linear models, returned as a structure with the fields described in this table.
Field in
FitInfo | Description |
|---|---|
Intercept | Intercept term
β0 for each
linear model, a 1-by-L
vector |
Lambda | Lambda parameters in ascending order, a
1-by-L
vector |
Alpha | Value of the Alpha parameter, a
scalar |
DF | Number of nonzero coefficients in B
for each value of Lambda, a
1-by-L
vector |
MSE | Mean squared error (MSE), a
1-by-L
vector |
PredictorNames | Value of the PredictorNames parameter,
stored as a cell array of character vectors |
UseCovariance | Logical value indicating whether the covariance matrix
was used in fitting. If the covariance was computed and
used, this field is true. Otherwise, this
field is false. |
If CV is a cvpartition object, the
FitInfo contains these additional fields.
Field in
FitInfo | Description |
|---|---|
SE | Standard error of MSE for each Lambda,
as calculated during cross-validation, a
1-by-L
vector |
LambdaMinMSE | Lambda value with the minimum MSE, a
scalar |
Lambda1SE | Largest Lambda value such that MSE is
within one standard error of the minimum MSE, a
scalar |
IndexMinMSE | Index of Lambda with the value
LambdaMinMSE, a scalar |
Index1SE | Index of Lambda with the value
Lambda1SE, a scalar |
More About
For a given value of λ, a nonnegative parameter,
lasso solves the problem
N is the number of observations.
yi is the response at observation i.
xi is data, a vector of length p at observation i.
λ is a nonnegative regularization parameter corresponding to one value of
Lambda.The parameters β0 and β are a scalar and a vector of length p, respectively.
As λ increases, the number of nonzero components of β decreases.
The lasso problem involves the L1 norm of β, as contrasted with the elastic net algorithm.
For α strictly between 0 and 1, and nonnegative λ, elastic net solves the problem
where
Elastic net is the same as lasso when α = 1. For
other values of α, the penalty term
Pα(β)
interpolates between the L1 norm of
β and the squared
L2 norm of β.
As α shrinks toward 0, elastic net approaches ridge regression.
Algorithms
lasso fits many values of λ
simultaneously by an efficient procedure named coordinate
descent, based on Friedman, Tibshirani, and Hastie [3]. The procedure has two main code paths depending on whether the fitting uses a
covariance matrix. You can affect this choice with the
UseCovariance name-value argument.
When lasso uses a covariance matrix to fit
N data points and D predictors, the
fitting has a rough computational complexity of D*D. Without a
covariance matrix, the computational complexity is roughly N*D.
So, typically, using a covariance matrix can be faster when N >
D, and the default 'auto' setting of the
UseCovariance argument makes this choice. Using a covariance
matrix causes lasso to subtract larger numbers than
otherwise, which can be less numerically stable. For details of the algorithmic
differences, see [3]. For one comparison of timing and accuracy
differences, see Use Correlation Matrix for Fitting Lasso.
When operating on tall arrays, lasso uses an algorithm based
on the Alternating Direction Method of Multipliers (ADMM) [5]. The notation used here is the same as in the reference paper. This method solves
problems of the form
Minimize
Subject to
Using this notation, the lasso regression problem is
Minimize
Subject to
Because the loss function is quadratic, the iterative updates performed by the algorithm amount to solving a linear system of equations with a single coefficient matrix but several right-hand sides. The updates performed by the algorithm during each iteration are
A is the dataset (a tall array), x contains the coefficients, ρ is the penalty parameter (augmented Lagrangian parameter), b is the response (a tall array), and S is the soft thresholding operator.
lasso solves the linear system using Cholesky factorization
because the coefficient matrix is symmetric and positive definite. Because does not change between iterations, the Cholesky factorization is
cached between iterations.
Even though A and b are tall arrays, they appear only in the terms and . The results of these two matrix multiplications are small enough to fit in memory, so they are precomputed and the iterative updates between iterations are performed entirely within memory.
References
[1] Tibshirani, R. “Regression Shrinkage and Selection via the Lasso.” Journal of the Royal Statistical Society. Series B, Vol. 58, No. 1, 1996, pp. 267–288.
[2] Zou, H., and T. Hastie. “Regularization and Variable Selection via the Elastic Net.” Journal of the Royal Statistical Society. Series B, Vol. 67, No. 2, 2005, pp. 301–320.
[3] Friedman, J., R. Tibshirani, and T. Hastie.
“Regularization Paths for Generalized Linear Models via Coordinate
Descent.” Journal of Statistical Software. Vol. 33, No. 1,
2010. https://www.jstatsoft.org/v33/i01
[4] Hastie, T., R. Tibshirani, and J. Friedman. The Elements of Statistical Learning. 2nd edition. New York: Springer, 2008.
[5] Boyd, S. “Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers.” Foundations and Trends in Machine Learning. Vol. 3, No. 1, 2010, pp. 1–122.
Extended Capabilities
This function supports tall arrays for out-of-memory data with some limitations.
With tall arrays,
lassouses an algorithm based on ADMM (Alternating Direction Method of Multipliers).No elastic net support. The
Alphaparameter is always 1.No cross-validation (
CVname-value argument) support, which includes the related name-value argumentMCReps.The output
FitInfodoes not contain the additional fields'SE','LambdaMinMSE','Lambda1SE','IndexMinMSE', and'Index1SE'.The
Optionsname-value argument is not supported because it does not contain options that apply to the ADMM algorithm. You can tune the ADMM algorithm using name-value arguments.Supported name-value arguments are:
LambdaLambdaRatioNumLambdaStandardizePredictorNamesRelTolWeights
Additional name-value arguments to control the ADMM algorithm are:
Rho— Augmented Lagrangian parameter, ρ. The default value is automatic selection.AbsTol— Absolute tolerance used to determine convergence. The default value is1e–4.MaxIter— Maximum number of iterations. The default value is1e4.B0— Initial values for the coefficients x. The default value is a vector of zeros.U0— Initial values of the scaled dual variable u. The default value is a vector of zeros.
For more information, see Tall Arrays.
To run in parallel, specify the Options name-value argument.
Set the UseParallel field of the options structure to
true using statset:
Options=statset(UseParallel=true)
Parallel computation is not used when CV is
"resubstitution". For more information about parallel
computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Version History
Introduced in R2011b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)