assignClusters
Description
idx = assignClusters(Mdl,X)X, using the incrementalKMeans
model Mdl. You cannot call assignClusters if
Mdl.NumPredictors is 0 or if all the values of
Mdl.Centroids are NaN. When you call
assignClusters, the software does not update
Mdl.
Examples
Create an incremental model for k-means clustering that has two clusters.
Mdl = incrementalKMeans(numClusters=2)
Mdl =
incrementalKMeans
IsWarm: 0
Metrics: [1×2 table]
NumClusters: 2
Centroids: [2×0 double]
Distance: "sqeuclidean"
Properties, Methods
Mdl is an incrementalKMeans model object. All its properties are read-only.
Load and Preprocess Data
Load the New York city housing data set.
load NYCHousing2015.matThe data set includes 10 variables with information on the sales of properties in New York City in 2015. Keep only the gross square footage and sale price predictors. Keep all records that have a gross square footage above 100 square feet and a sales price above $1000.
data = NYCHousing2015(:,{'GROSSSQUAREFEET','SALEPRICE'});
data = data((data.GROSSSQUAREFEET > 100 & data.SALEPRICE > 1000),:);Convert the tabular data into a matrix that contains the logarithm of both predictors.
X = table2array(log10(data));
Randomly shuffle the order of the records.
rng(0,"twister"); % For reproducibility X = X(randperm(size(X,1)),:);
Fit and Plot Incremental Model
Fit the incremental model Mdl to the data by using the fit function. To simulate a data stream, fit the model in chunks of 500 records at a time. At each iteration:
Process 500 observations.
Overwrite the previous incremental model with a new one fitted to the incoming records.
Update the performance metrics for the model. The default metric for
MdlisSimplifiedSilhouette.Store the cumulative and window metrics to see how they evolve during incremental learning.
Compute the cluster assignments of all records seen so far, according to the current model.
Plot all records seen so far, and color each record by its cluster assignment.
Plot the current centroid location of each cluster.
In this workflow, the updateMetrics function provides information about the model's clustering performance after it is fit to the incoming data chunk. In other workflows, you might want to evaluate a clustering model's performance on unseen data. In such cases, you can call updateMetrics prior to calling the incremental fit function.
% Initialize plot properties hold on h1 = scatter(NaN,NaN,0.3); h2 = plot(NaN,NaN,Marker="o", ... MarkerFaceColor="k",MarkerEdgeColor="k"); h3 = plot(NaN,NaN,Marker="^", ... MarkerFaceColor="b",MarkerEdgeColor="b"); colormap(gca,"prism") pbaspect([1,1,1]) xlim([min(X(:,1)),max(X(:,1))]); ylim([min(X(:,2)),max(X(:,2))]); xlabel("log Gross Square Footage"); ylabel("log Sales Price in Dollars") % Incremental fitting and plotting n = numel(X(:,1)); numObsPerChunk = 500; nchunk = floor(n/numObsPerChunk); sil = array2table(zeros(nchunk,2),VariableNames=["Cumulative" "Window"]); for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; Mdl = fit(Mdl,X(idx,:)); Mdl = updateMetrics(Mdl,X(idx,:)); sil{j,:} = Mdl.Metrics{'SimplifiedSilhouette',:}; indices = assignClusters(Mdl,X(1:iend,:)); title("Iteration " + num2str(j)) set(h1,XData=X(1:iend,1),YData=X(1:iend,2),CData=indices); set(h2,Marker="none") % Erase previous centroid markers set(h3,Marker="none") set(h2,XData=Mdl.Centroids(1,1),YData=Mdl.Centroids(1,2),Marker="o") set(h3,Xdata=Mdl.Centroids(2,1),YData=Mdl.Centroids(2,2),Marker="^") pause(0.5); end
Warning: Hardware-accelerated graphics is unavailable. Displaying fewer markers to preserve interactivity.
hold off
To view the animated figure, you can run the example, or open the animated gif below in your web browser.
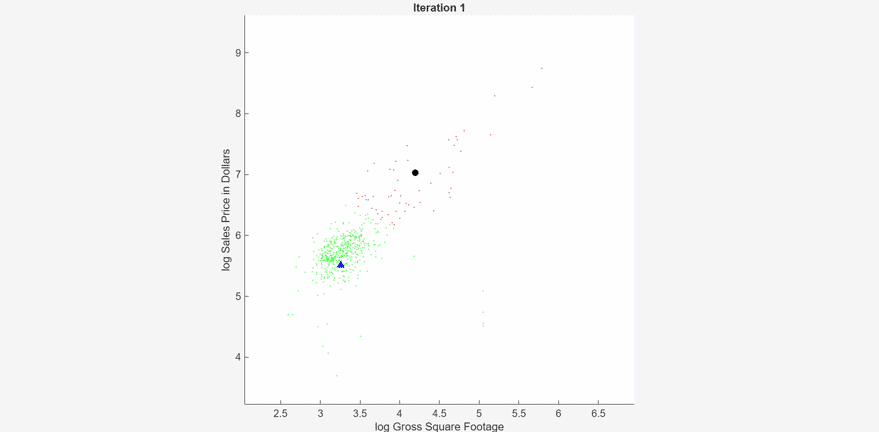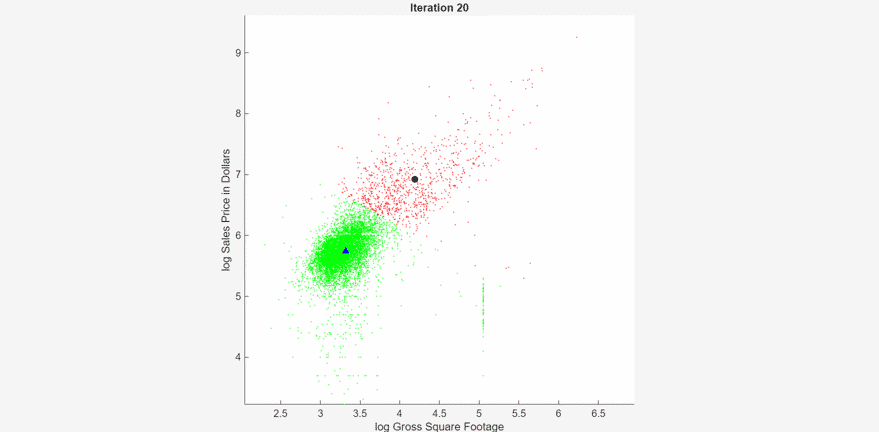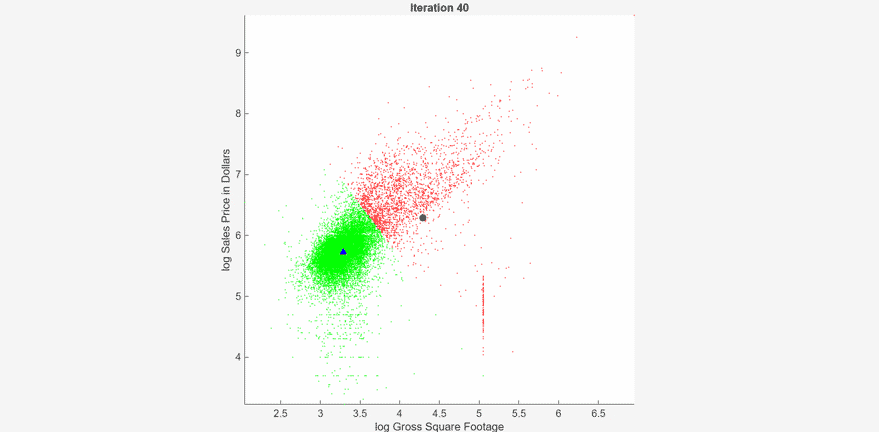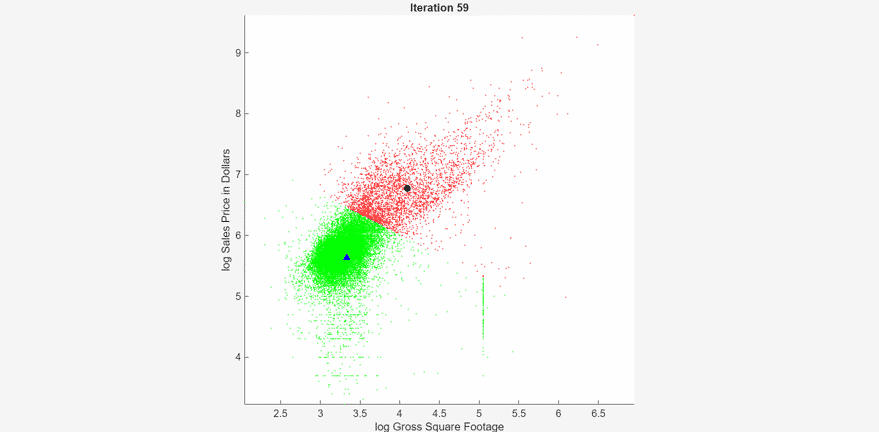
At each iteration, the animated plot displays all the observations processed so far as small circles, and colors them according to the cluster assignments of the current model. The black circle indicates the centroid position of cluster 1, and the blue triangle indicates the centroid position of cluster 2.
Plot the window and cumulative metrics values at each iteration.
h4 = plot(sil.Variables); xlabel("Iteration") ylabel("Performance Metric") xline(Mdl.WarmupPeriod/numObsPerChunk,'g-.') legend(h4,sil.Properties.VariableNames,Location="southeast")

The updateMetrics function calculates the performance metrics after the end of the warm-up period. The performance metrics rise rapidly from an initial value of 0.81 and approach a value of approximately 0.88 after 10 iterations.
Input Arguments
Incremental k-means clustering model, specified as an incrementalKMeans model object. You can create Mdl by
calling incrementalKMeans directly.
Chunk of predictor data, specified as an
n-by-Mdl.NumPredictors numeric matrix. The rows
of X correspond to observations, and the columns correspond to
predictor variables. If a row of X contains a missing value, the
corresponding values of idx and D for that row
are NaN.
Note
assignClusters supports
only numeric input predictor data. If your input data includes categorical data, you
must prepare an encoded version of the categorical data. Use dummyvar to convert each categorical variable to a numeric matrix of
dummy variables. Then, concatenate all dummy variable matrices and any other numeric
predictors. For more details, see Dummy Variables.
Data Types: single | double
Output Arguments
Cluster indices, returned as a size(X,1)-by-1 vector of integers.
If a row of X contains a missing value, the corresponding value of
idx is NaN. assignClusters
does not return indices of clusters whose corresponding Centroids
values are NaN.
Cluster distances, returned as a
size(X,1)-by-Mdl.NumClusters numeric matrix.
Each row in D contains the distance of the corresponding
observation in X from each cluster centroid, according to the
distance metric in Mdl.Distance. If a cluster has no observations
assigned to it, or its corresponding Centroids values are
NaN, the distance value for all observations to that cluster is
NaN. If a row of X contains a missing value,
the corresponding row of D contains all NaN
values.
Version History
Introduced in R2025a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)